1HTR
 
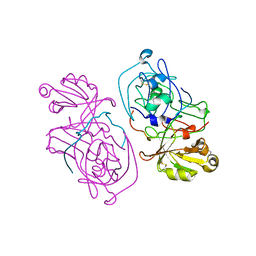 | |
1C2X
 
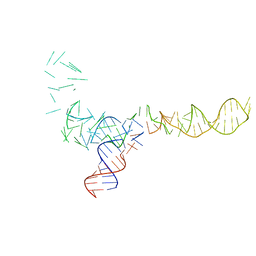 | |
1QL4
 
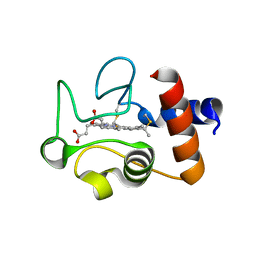 | | Structure of the soluble domain of cytochrome c552 from Paracoccus denitrificans in the oxidised state | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Harrenga, A, Reincke, B, Rueterjans, H, Ludwig, B, Michel, H. | | Deposit date: | 1999-08-20 | | Release date: | 2000-02-03 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Soluble Domain of Cytochrome C552 from Paracoccus Denitrificans in the Oxidized and Reduced States
J.Mol.Biol., 295, 2000
|
|
2G44
 
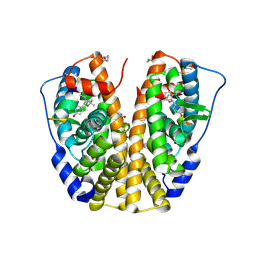 | | Human Estrogen Receptor Alpha Ligand-Binding Domain In Complex With OBCP-1M-G and A Glucocorticoid Receptor Interacting Protein 1 NR Box II Peptide | | Descriptor: | 4-[(1S,2R,5S)-4,4,8-TRIMETHYL-3-OXABICYCLO[3.3.1]NON-7-EN-2-YL]PHENOL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Greene, G.L. | | Deposit date: | 2006-02-21 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and characterization of novel estrogen
receptor agonist ligands and development of biochips for
nuclear receptor drug discovery
Thesis, 2006
|
|
6NJL
 
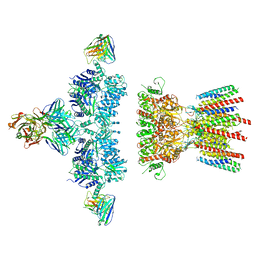 | |
7BU6
 
 | | Structure of human beta1 adrenergic receptor bound to norepinephrine and nanobody 6B9 | | Descriptor: | (2S)-2,3-dihydroxypropyl octanoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHOLESTEROL, ... | | Authors: | Xu, X, Kaindl, J, Clark, M, Hubner, H, Hirata, K, Sunahara, R, Gmeiner, P, Kobilka, B.K, Liu, X. | | Deposit date: | 2020-04-04 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding pathway determines norepinephrine selectivity for the human beta 1 AR over beta 2 AR.
Cell Res., 31, 2021
|
|
6O0X
 
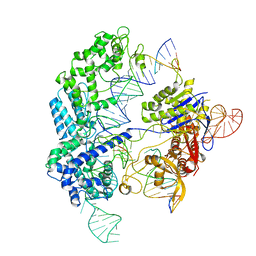 | | Conformational states of Cas9-sgRNA-DNA ternary complex in the presence of magnesium | | Descriptor: | 3' product of target strand DNA, 5' product of target strand DNA, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Zhu, X, Clarke, R, Puppala, A.K, Chittori, S, Merk, A, Merrill, B.J, Simonovic, M, Subramaniam, S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5AAV
 
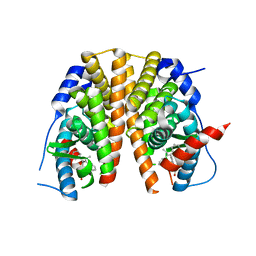 | | Optimization of a novel binding motif to to (E)-3-(3,5-difluoro-4-((1R,3R)-2-(2-fluoro-2-methylpropyl)-3-methyl-2,3,4,9-tetrahydro-1H- pyrido(3,4-b)indol-1-yl)phenyl)acrylic acid (AZD9496), a potent and orally bioavailable selective estrogen receptor downregulator and antagonist | | Descriptor: | (2E)-3-{4-[(1E)-1,2-DIPHENYLBUT-1-ENYL]PHENYL}ACRYLIC ACID, ESTROGEN RECEPTOR | | Authors: | Norman, R.A, Bradbury, R.H, de Almeida, C, Andrews, D.M, Ballard, P, Buttar, D, Callis, R.J, Currie, G.S, Curwen, J.O, Davies, C.D, de Savi, C, Donald, C.S, Feron, L.J.L, Glossop, S.C, Hayter, B.R, Karoutchi, G, Lamont, S.G, MacFaul, P, Moss, T, Pearson, S.E, Rabow, A.A, Tonge, M, Walker, G.E, Weir, H.M, Wilson, Z. | | Deposit date: | 2015-07-29 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Optimization of a Novel Binding Motif to (E)-3-(3,5-Difluoro-4-((1R,3R)-2-(2-Fluoro-2-Methylpropyl)-3-Methyl-2, 3,4,9-Tetrahydro-1H-Pyrido[3,4-B]Indol-1-Yl)Phenyl)Acrylic Acid (Azd9496), a Potent and Orally Bioavailable Selective Estrogen Receptor Downregulator and Antagonist.
J.Med.Chem., 58, 2015
|
|
5AAU
 
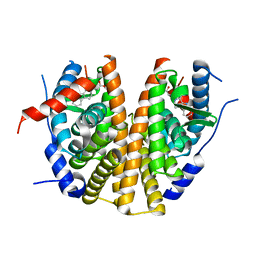 | | Optimization of a novel binding motif to to (E)-3-(3,5-difluoro-4-((1R,3R)-2-(2-fluoro-2-methylpropyl)-3-methyl-2,3,4,9-tetrahydro-1H- pyrido(3,4-b)indol-1-yl)phenyl)acrylic acid (AZD9496), a potent and orally bioavailable selective estrogen receptor downregulator and antagonist | | Descriptor: | 3-(1-(4-Chlorophenyl)-3,4-dihydro-1H-pyrido(3,4-b)indol-2(9H)-yl)propanoic acid, ESTROGEN RECEPTOR | | Authors: | Norman, R.A, Bradbury, R.H, de Almeida, C, Andrews, D.M, Ballard, P, Buttar, D, Callis, R.J, Currie, G.S, Curwen, J.O, Davies, C.D, de Savi, C, Donald, C.S, Feron, L.J.L, Glossop, S.C, Hayter, B.R, Karoutchi, G, Lamont, S.G, MacFaul, P, Moss, T, Pearson, S.E, Rabow, A.A, Tonge, M, Walker, G.E, Weir, H.M, Wilson, Z. | | Deposit date: | 2015-07-28 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of a Novel Binding Motif to (E)-3-(3,5-Difluoro-4-((1R,3R)-2-(2-Fluoro-2-Methylpropyl)-3-Methyl-2, 3,4,9-Tetrahydro-1H-Pyrido[3,4-B]Indol-1-Yl)Phenyl)Acrylic Acid (Azd9496), a Potent and Orally Bioavailable Selective Estrogen Receptor Downregulator and Antagonist.
J.Med.Chem., 58, 2015
|
|
5ACC
 
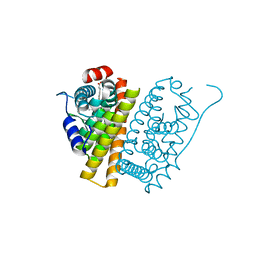 | | A Novel Oral Selective Estrogen Receptor Down-regulator, AZD9496, drives Tumour Growth Inhibition in Estrogen Receptor positive and ESR1 Mutant Models | | Descriptor: | (E)-3-(3,5-DIFLUORO-4-((1R,3R)-2-(2-FLUORO-2- METHYLPROPYL)-3-METHYL-2,3,4,9-TETRAHYDRO-1H-PYRIDO(3,4-B)INDOL-1-YL)PHENYL)ACRYLIC ACID, ESTROGEN RECEPTOR | | Authors: | Norman, R.A, Weir, H.M, Bradbury, R.H, Lawson, M, Rabow, A.A, Buttar, D, Callis, R.J, Curwen, J.O, de Almeida, C, Ballard, P, Hulse, M, Donald, C.S, Feron, L.J.L, Gingell, H, Karoutchi, G, MacFaul, P, Moss, T, Pearson, S.E, Tonge, M, Davies, G, Walker, G.E, Wilson, Z, Rowlinson, R, Powell, S, Hemsley, P, Linney, E, Campbell, H, Ghazoui, Z, Sadler, C, Richmond, G, Pazolli, E, Mazzola, A.M, DCruz, C, De Savi, C. | | Deposit date: | 2015-08-15 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Optimization of a Novel Binding Motif to (E)-3-(3,5-Difluoro-4-((1R,3R)-2-(2-Fluoro-2-Methylpropyl)-3-Methyl-2, 3,4,9-Tetrahydro-1H-Pyrido[3,4-B]Indol-1-Yl)Phenyl)Acrylic Acid (Azd9496), a Potent and Orally Bioavailable Selective Estrogen Receptor Downregulator and Antagonist.
J.Med.Chem., 58, 2015
|
|
1BOU
 
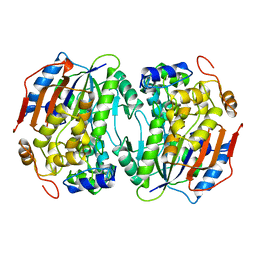 | | THREE-DIMENSIONAL STRUCTURE OF LIGAB | | Descriptor: | 4,5-DIOXYGENASE ALPHA CHAIN, 4,5-DIOXYGENASE BETA CHAIN, FE (III) ION | | Authors: | Sugimoto, K, Senda, T, Fukuda, M, Mitsui, Y. | | Deposit date: | 1998-08-06 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an aromatic ring opening dioxygenase LigAB, a protocatechuate 4,5-dioxygenase, under aerobic conditions.
Structure, 7, 1999
|
|
6NBS
 
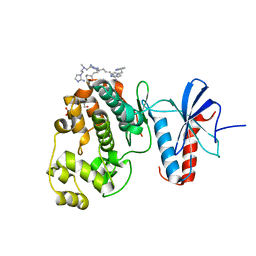 | | WT ERK2 with compound 2507-8 | | Descriptor: | (5S)-5-benzyl-4,5-dihydro-1H-imidazol-2-amine, GLYCEROL, Mitogen-activated protein kinase 1, ... | | Authors: | Sammons, R.M, Perry, N.A, Cho, E.J, Kaoud, T.S, Zamora-Olivares, D.P, Piserchio, A, Houghten, R.A, Giulianotti, M, Li, Y, Debevec, G, Gurevich, V.V, Ghose, R, Iverson, T.M, Dalby, K.N. | | Deposit date: | 2018-12-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Class of Common Docking Domain Inhibitors That Prevent ERK2 Activation and Substrate Phosphorylation.
Acs Chem.Biol., 14, 2019
|
|
7F3F
 
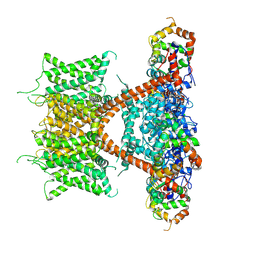 | | CryoEM structure of human Kv4.2-KChIP1 complex | | Descriptor: | Isoform 2 of Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-06-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
1THP
 
 | | STRUCTURE OF HUMAN ALPHA-THROMBIN Y225P MUTANT BOUND TO D-PHE-PRO-ARG-CHLOROMETHYLKETONE | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, PROTEIN (THROMBIN HEAVY CHAIN), PROTEIN (THROMBIN LIGHT CHAIN) | | Authors: | Caccia, S, Futterer, K, Di Cera, E, Waksman, G. | | Deposit date: | 1999-01-26 | | Release date: | 1999-03-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected crucial role of residue 225 in serine proteases.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7F0J
 
 | | CryoEM structure of human Kv4.2 | | Descriptor: | Potassium voltage-gated channel subfamily D member 2, ZINC ION | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-06-04 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
1CMO
 
 | | IMMUNOGLOBULIN MOTIF DNA-RECOGNITION AND HETERODIMERIZATION FOR THE PEBP2/CBF RUNT-DOMAIN | | Descriptor: | POLYOMAVIRUS ENHANCER BINDING PROTEIN 2 | | Authors: | Nagata, T, Gupta, V, Sorce, D, Kim, W.Y, Sali, A, Chait, B.T, Shigesada, K, Ito, Y, Werner, M.H. | | Deposit date: | 1999-05-11 | | Release date: | 2000-01-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain.
Nat.Struct.Biol., 6, 1999
|
|
6NJN
 
 | |
1CSJ
 
 | | CRYSTAL STRUCTURE OF THE RNA-DEPENDENT RNA POLYMERASE OF HEPATITIS C VIRUS | | Descriptor: | HEPATITIS C VIRUS RNA POLYMERASE (NS5B) | | Authors: | Bressanelli, S, Tomei, L, Roussel, A, Incitti, I, Vitale, R.L, Mathieu, M, De Francesco, R, Rey, F.A. | | Deposit date: | 1999-08-18 | | Release date: | 1999-11-08 | | Last modified: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1JT8
 
 | | ARCHAEAL INITIATION FACTOR-1A, AIF-1A | | Descriptor: | PROBABLE TRANSLATION INITIATION FACTOR 1A | | Authors: | Hoffman, D.W, Li, W. | | Deposit date: | 2001-08-20 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of translation initiation factor aIF-1A from the archaeon Methanococcus jannaschii determined by NMR spectroscopy
Protein Sci., 10, 2001
|
|
2D1X
 
 | | The crystal structure of the cortactin-SH3 domain and AMAP1-peptide complex | | Descriptor: | SULFATE ION, cortactin isoform a, proline rich region from development and differentiation enhancing factor 1 | | Authors: | Hashimoto, S, Hirose, M, Hashimoto, A, Morishige, M, Yamada, A, Hosaka, H, Akagi, K, Ogawa, E, Oneyama, C, Agatsuma, T, Okada, M, Kobayashi, H, Wada, H, Nakano, H, Ikegami, T, Nakagawa, A, Sabe, H. | | Deposit date: | 2005-09-01 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting AMAP1 and cortactin binding bearing an atypical src homology 3/proline interface for prevention of breast cancer invasion and metastasis.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1UA2
 
 | | Crystal Structure of Human CDK7 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein kinase 7 | | Authors: | Lolli, G, Lowe, E.D, Brown, N.R, Johnson, L.N. | | Deposit date: | 2004-08-11 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The Crystal Structure of Human CDK7 and Its Protein Recognition Properties
Structure, 12, 2004
|
|
7SYO
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head open. Structure 9(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SML
 
 | | Crystal Structure of L-GALACTONO-1,4-LACTONE DEHYDROGENASE de Myrciaria dubia | | Descriptor: | L-GALACTONO-1,4-LACTONE DEHYDROGENASE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2021-10-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit Camu-Camu.
J.Exp.Bot., 2024
|
|
6NWP
 
 | | Chronic traumatic encephalopathy Type I Tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Falcon, B, Zivanov, J, Zhang, W, Murzin, A.G, Garringer, H.J, Vidal, R, Crowther, R.A, Newell, K.L, Ghetti, B, Goedert, M, Scheres, H.W. | | Deposit date: | 2019-02-07 | | Release date: | 2019-03-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules.
Nature, 568, 2019
|
|
6TET
 
 | | The structure of CYP121 in complex with inhibitor L21 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(~{E})-3-[4-(4-fluorophenyl)phenyl]prop-2-enyl]imidazole, Mycocyclosin synthase, ... | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2019-11-12 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49986887 Å) | | Cite: | Structure-Activity Relationship and Mode-Of-Action Studies Highlight 1-(4-Biphenylylmethyl)-1H-imidazole-Derived Small Molecules as Potent CYP121 Inhibitors.
Chemmedchem, 16, 2021
|
|
