5OB4
 
 | |
6BNQ
 
 | |
2OEM
 
 | | Crystal structure of a rubisco-like protein from Geobacillus kaustophilus liganded with Mg2+ and 2,3-diketohexane 1-phosphate | | Descriptor: | (1Z)-2-HYDROXY-3-OXOHEX-1-EN-1-YL DIHYDROGEN PHOSPHATE, 2,3-diketo-5-methylthiopentyl-1-phosphate enolase, MAGNESIUM ION | | Authors: | Fedorov, A.A, Imker, H.J, Fedorov, E.V, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-12-30 | | Release date: | 2007-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanistic Diversity in the RuBisCO Superfamily: The "Enolase" in the Methionine Salvage Pathway in Geobacillus kaustophilus.
Biochemistry, 46, 2007
|
|
3UPU
 
 | | Crystal structure of the T4 Phage SF1B Helicase Dda | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*T)-3', ATP-dependent DNA helicase dda | | Authors: | He, X, Yun, M.K, Pemble IV, C.W, Kreuzer, K.N, Raney, K.D, White, S.W. | | Deposit date: | 2011-11-18 | | Release date: | 2012-06-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | The T4 Phage SF1B Helicase Dda Is Structurally Optimized to Perform DNA Strand Separation.
Structure, 20, 2012
|
|
2OEJ
 
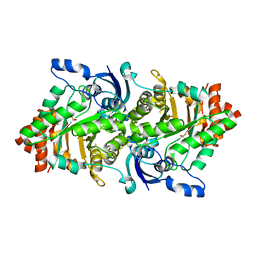 | | Crystal structure of a rubisco-like protein from Geobacillus kaustophilus (tetramutant form), liganded with phosphate ions | | Descriptor: | 2,3-diketo-5-methylthiopentyl-1-phosphate enolase, PHOSPHATE ION | | Authors: | Fedorov, A.A, Imker, H.J, Fedorov, E.V, Almo, S.C, Gerlt, J.A. | | Deposit date: | 2006-12-30 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanistic Diversity in the RuBisCO Superfamily: The "Enolase" in the Methionine Salvage Pathway in Geobacillus kaustophilus.
Biochemistry, 46, 2007
|
|
4DYZ
 
 | |
4DZ0
 
 | |
1DBI
 
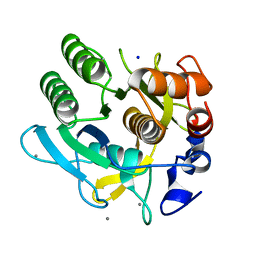 | | CRYSTAL STRUCTURE OF A THERMOSTABLE SERINE PROTEASE | | Descriptor: | AK.1 SERINE PROTEASE, CALCIUM ION, SODIUM ION | | Authors: | Smith, C.A, Toogood, H.S, Baker, H.M, Daniel, R.M, Baker, E.N. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-18 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Calcium-mediated thermostability in the subtilisin superfamily: the crystal structure of Bacillus Ak.1 protease at 1.8 A resolution.
J.Mol.Biol., 294, 1999
|
|
2Q1A
 
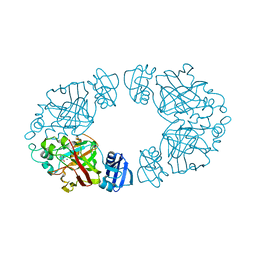 | | 2-keto-3-deoxy-D-arabinonate dehydratase complexed with magnesium and 2-oxobutyrate | | Descriptor: | 2-KETOBUTYRIC ACID, 2-keto-3-deoxy-D-arabinonate dehydratase, MAGNESIUM ION | | Authors: | Barends, T, Brouns, S, Worm, P, Akerboom, J, Turnbull, A, Salmon, L. | | Deposit date: | 2007-05-24 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily.
J.Mol.Biol., 379, 2008
|
|
1UKC
 
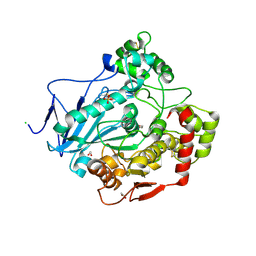 | | Crystal Structure of Aspergillus niger EstA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bourne, Y, Hasper, A.A, Chahinian, H, Juin, M, De Graaff, L.H, Marchot, P. | | Deposit date: | 2003-08-19 | | Release date: | 2004-07-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aspergillus niger protein EstA defines a new class of fungal esterases within the alpha/beta hydrolase fold superfamily of proteins
STRUCTURE, 12, 2004
|
|
2Q18
 
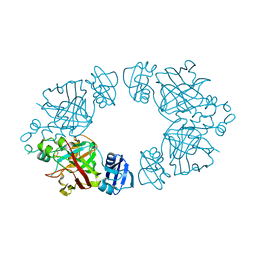 | | 2-keto-3-deoxy-D-arabinonate dehydratase | | Descriptor: | 2-keto-3-deoxy-D-arabinonate dehydratase, PHOSPHATE ION | | Authors: | Barends, T, Brouns, S, Worm, P, Akerboom, J, Turnbull, A, Salmon, L. | | Deposit date: | 2007-05-24 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily.
J.Mol.Biol., 379, 2008
|
|
1LHP
 
 | |
4LRU
 
 | | Crystal structure of glyoxalase III (Orf 19.251) from Candida albicans | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glyoxalase III (glutathione-independent) | | Authors: | Hasim, S, Hussin, N.A, Nickerson, K.W, Wilson, M.A. | | Deposit date: | 2013-07-20 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Glutathione-independent Glyoxalase of the DJ-1 Superfamily Plays an Important Role in Managing Metabolically Generated Methylglyoxal in Candida albicans.
J.Biol.Chem., 289, 2014
|
|
3U1D
 
 | | The structure of a protein with a GntR superfamily winged-helix-turn-helix domain from Halomicrobium mukohataei. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Cuff, M.E, Bigelow, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-07 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a protein with a GntR superfamily winged-helix-turn-helix domain from Halomicrobium mukohataei.
TO BE PUBLISHED
|
|
1LHR
 
 | | Crystal Structure of Pyridoxal Kinase complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, POTASSIUM ION, Pyridoxal kinase, ... | | Authors: | Liang, D.C, Jiang, T, Li, M.H. | | Deposit date: | 2002-04-17 | | Release date: | 2003-02-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of brain pyridoxal kinase, a novel member of the ribokinase superfamily
J.BIOL.CHEM., 277, 2002
|
|
4FYP
 
 | | Crystal Structure of Plant Vegetative Storage Protein | | Descriptor: | MAGNESIUM ION, Vegetative storage protein 1 | | Authors: | Chen, Y, Wei, J, Wang, M, Gong, W, Zhang, M. | | Deposit date: | 2012-07-05 | | Release date: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of Arabidopsis VSP1 reveals the plant class C-like phosphatase structure of the DDDD superfamily of phosphohydrolases
Plos One, 7, 2012
|
|
8DLE
 
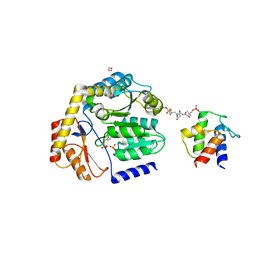 | | Crosslinked Crystal Structure of the 8-amino-7-oxonanoate synthase, BioF, and Benzene Sulfonyl Fluoride-crypto Acyl Carrier Protein, BSF-ACP | | Descriptor: | 1,2-ETHANEDIOL, 8-amino-7-oxononanoate synthase, Acyl carrier protein, ... | | Authors: | Chen, A, Davis, T.D, Louie, G.V, Bowman, M.E, Noel, J.P, Burkart, M.D. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Visualizing the Interface of Biotin and Fatty Acid Biosynthesis through SuFEx Probes.
J.Am.Chem.Soc., 146, 2024
|
|
7PA5
 
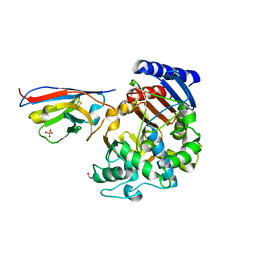 | | Complex between the beta-lactamase CMY-2 with an inhibitory nanobody | | Descriptor: | Beta-lactamase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Frederic Cawez, F.C, Frederic Kerff, F.K, Moreno Galleni, M.G, Raphael Herman, R.H. | | Deposit date: | 2021-07-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.184 Å) | | Cite: | Development of Nanobodies as Theranostic Agents against CMY-2-Like Class C beta-Lactamases.
Antimicrob.Agents Chemother., 67, 2023
|
|
7PCU
 
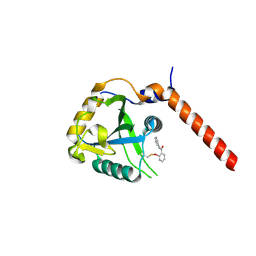 | | Crystal structure of YTHDF1 YTH domain in complex with ebselen | | Descriptor: | N-phenyl-2-selanylbenzamide, YTH domain-containing family protein 1 | | Authors: | Dalle Vedove, A, Cazzanelli, G, Quattrone, A, Provenzani, A, Lolli, G. | | Deposit date: | 2021-08-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small-Molecule Ebselen Binds to YTHDF Proteins Interfering with the Recognition of N 6 -Methyladenosine-Modified RNAs.
Acs Pharmacol Transl Sci, 5, 2022
|
|
2W3P
 
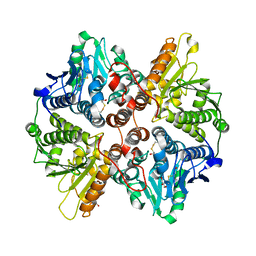 | | BoxC crystal structure | | Descriptor: | BENZOYL-COA-DIHYDRODIOL LYASE, BETA-MERCAPTOETHANOL, GLYCEROL | | Authors: | Bains, J, Boulanger, M.J. | | Deposit date: | 2008-11-13 | | Release date: | 2009-04-14 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Biophysical Characterization of Boxc from Burkholderia Xenovorans Lb400: A Novel Ring-Cleaving Enzyme in the Crotonase Superfamily.
J.Biol.Chem., 284, 2009
|
|
8EOA
 
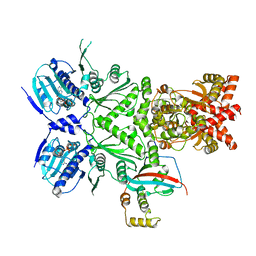 | | Cryo-EM structure of human HSP90B-AIPL1 complex | | Descriptor: | Aryl-hydrocarbon-interacting protein-like 1, Heat shock protein HSP 90-beta, MAGNESIUM ION, ... | | Authors: | Srivastava, D, Artemyev, N.O. | | Deposit date: | 2022-10-02 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Unique interface and dynamics of the complex of HSP90 with a specialized cochaperone AIPL1.
Structure, 31, 2023
|
|
8EOB
 
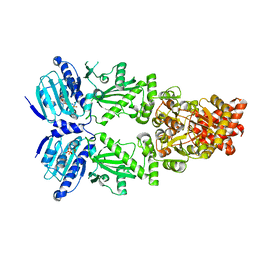 | | Cryo-EM structure of human HSP90B in the closed state | | Descriptor: | Heat shock protein HSP 90-beta, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Srivastava, D, Artemyev, N.O. | | Deposit date: | 2022-10-02 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unique interface and dynamics of the complex of HSP90 with a specialized cochaperone AIPL1.
Structure, 31, 2023
|
|
2W8M
 
 | |
1Z0R
 
 | | Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transcription-State Regulator AbrB | | Descriptor: | Transition state regulatory protein abrB | | Authors: | Bobay, B.G, Andreeva, A, Mueller, G.A, Cavanagh, J, Murzin, A.G. | | Deposit date: | 2005-03-02 | | Release date: | 2005-03-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Revised structure of the AbrB N-terminal domain unifies a diverse superfamily of putative DNA-binding proteins.
Febs Lett., 579, 2005
|
|
2A7K
 
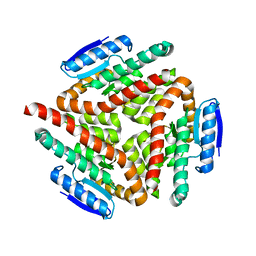 | | carboxymethylproline synthase (CarB) from pectobacterium carotovora, apo enzyme | | Descriptor: | CarB | | Authors: | Sleeman, M.C, Sorensen, J.L, Batchelar, E.T, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2005-07-05 | | Release date: | 2005-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural and Mechanistic Studies on Carboxymethylproline Synthase (CarB), a Unique Member of the Crotonase Superfamily Catalyzing the First Step in Carbapenem Biosynthesis.
J.Biol.Chem., 280, 2005
|
|
