6ZYA
 
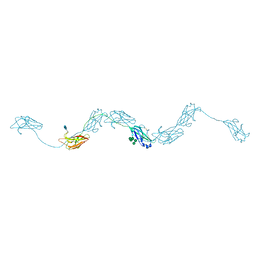 | | Extended human uromodulin filament core at 3.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
6ZOX
 
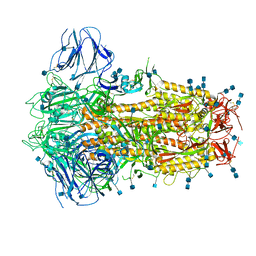 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x2 disulphide-bond mutant, G413C, V987C, single Arg S1/S2 cleavage site) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZYM
 
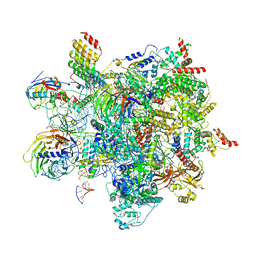 | | Human C Complex Spliceosome - High-resolution CORE | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Corepressor interacting with RBPJ 1, ... | | Authors: | Bertram, K, Kastner, B. | | Deposit date: | 2020-08-02 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insights into the Roles of Metazoan-Specific Splicing Factors in the Human Step 1 Spliceosome.
Mol.Cell, 80, 2020
|
|
3KVO
 
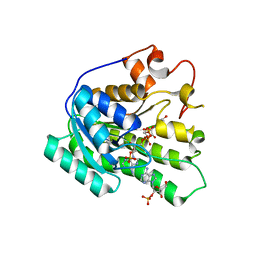 | | Crystal structure of the catalytic domain of human Hydroxysteroid dehydrogenase like 2 (HSDL2) | | Descriptor: | Hydroxysteroid dehydrogenase-like protein 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ugochukwu, E, Bhatia, C, Huang, J, Pilka, E, Muniz, J.R.C, Pike, A.C.W, Krojer, T, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Verdin, E.M, Oppermann, U, Kavanagh, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-30 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the catalytic domain of human Hydroxysteroid dehydrogenase like 2 (HSDL2)
To be Published
|
|
3KXD
 
 | |
3L3O
 
 | |
3KPK
 
 | | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans, C160A mutant | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, ... | | Authors: | Cherney, M.M, Zhang, Y, Solomonson, M, Weiner, J.H, James, M.N.G. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans: insights into sulfidotrophic respiration and detoxification.
J.Mol.Biol., 398, 2010
|
|
3L0K
 
 | |
3KPN
 
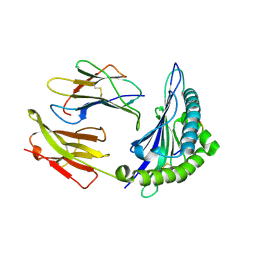 | | Crystal Structure of HLA B*4403 in complex with EEYLQAFTY a self peptide from the ABCD3 protein | | Descriptor: | Beta-2-microglobulin, EEYLQAFTY, self peptide from the ATP binding cassette protein ABCD3, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3KS0
 
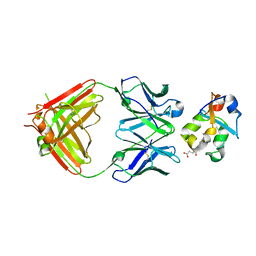 | | Crystal structure of the heme domain of flavocytochrome b2 in complex with Fab B2B4 | | Descriptor: | Cytochrome b2, mitochondrial, Fragment Antigen Binding B2B4, ... | | Authors: | Golinelli-Pimpaneau, B, Lederer, F, Le, K.H.D. | | Deposit date: | 2009-11-20 | | Release date: | 2010-05-26 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for the functional importance of the heme domain mobility in flavocytochrome b2.
J.Mol.Biol., 400, 2010
|
|
3L0N
 
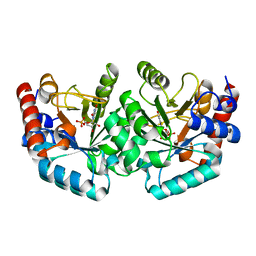 | | Human orotidyl-5'-monophosphate decarboxylase in complex with 6-mercapto-UMP | | Descriptor: | 6-sulfanyluridine-5'-phosphate, Uridine 5'-monophosphate synthase | | Authors: | Heinrich, D, Wittmann, J, Diederichsen, U, Rudolph, M. | | Deposit date: | 2009-12-10 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase
Chemistry, 15, 2009
|
|
8AP5
 
 | |
7PEB
 
 | |
7PEC
 
 | |
7PEA
 
 | |
8B6X
 
 | |
7PBW
 
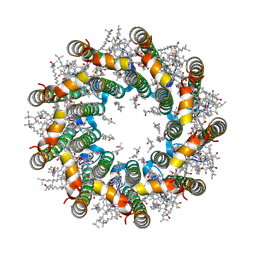 | | Cryo-EM structure of light harvesting complex 2 from Rba. sphaeroides. | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Qian, P, Swainsbury, D.J.K, Croll, T.I, Castro-Hartmann, P, Sader, K, Divitini, G, Hunter, C.N. | | Deposit date: | 2021-08-02 | | Release date: | 2021-11-24 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Cryo-EM Structure of the Rhodobacter sphaeroides Light-Harvesting 2 Complex at 2.1 angstrom.
Biochemistry, 60, 2021
|
|
8B6Y
 
 | |
7PTT
 
 | |
7PTP
 
 | |
7MPI
 
 | | Stm1 bound vacant 80S structure isolated from cbf5-D95A | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-11 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
8BDA
 
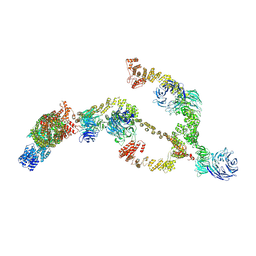 | | IFTA complex in anterograde intraflagellar transport trains (Chlamydomonas reinhardtii) | | Descriptor: | Intraflagellar transport particle protein 140, Intraflagellar transport protein 121, Intraflagellar transport protein 122 homolog, ... | | Authors: | Lacey, S.E, Foster, H.E, Pigino, G. | | Deposit date: | 2022-10-18 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (20.700001 Å) | | Cite: | The molecular structure of IFT-A and IFT-B in anterograde intraflagellar transport trains.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6BG9
 
 | |
7PHM
 
 | |
7RFA
 
 | |
