2ZVN
 
 | | NEMO CoZi domain incomplex with diubiquitin in P212121 space group | | Descriptor: | NF-kappa-B essential modulator, UBC protein | | Authors: | Rahighi, S, Ikeda, F, Kawasaki, M, Akutsu, M, Suzuki, N, Kato, R, Kensche, T, Uejima, T, Bloor, S, Komander, D, Randow, F, Wakatsuki, S, Dikic, I. | | Deposit date: | 2008-11-12 | | Release date: | 2009-03-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation
Cell(Cambridge,Mass.), 136, 2009
|
|
4M8E
 
 | | CRYSTAL STRUCTURE OF HUMAN RETINOID X RECEPTOR ALPHA-LIGAND BINDING DOMAIN COMPLEX WITH (S) 4-Methyl 9cUAB30 COACTIVATOR PEPTIDE GRIP-1 | | Descriptor: | (3E,6Z,8E)-3,7-dimethyl-8-[(4S)-4-methyl-3,4-dihydronaphthalen-1(2H)-ylidene]octa-3,6-dienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Smith, C.D, Muccio, D.D. | | Deposit date: | 2013-08-13 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Methyl-substituted conformationally constrained rexinoid agonists for the retinoid X receptors demonstrate improved efficacy for cancer therapy and prevention.
Bioorg.Med.Chem., 22, 2014
|
|
3N33
 
 | | Crystal structure of the N-terminal beta-aminopeptidase BapA in complex with pefabloc SC (AEBSF) | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-peptidyl aminopeptidase, ... | | Authors: | Merz, T, Heck, T, Geueke, B, Kohler, H.-P, Gruetter, M.G. | | Deposit date: | 2010-05-19 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Autoproteolytic and catalytic mechanisms for the beta-aminopeptidase BapA--a member of the Ntn hydrolase family.
Structure, 20, 2012
|
|
8BP2
 
 | | 2.8A STRUCTURE OF ZOLIFLODACIN WITH S.AUREUS DNA GYRASE AND DNA | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (20-MER), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Morgan, H, Warren, A.J. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A 2.8 angstrom Structure of Zoliflodacin in a DNA Cleavage Complex with Staphylococcus aureus DNA Gyrase.
Int J Mol Sci, 24, 2023
|
|
4JTL
 
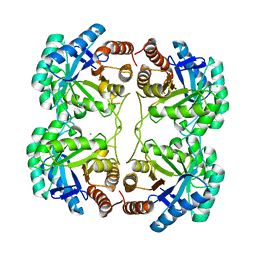 | | Crystal structure of F139G mutant of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION | | Authors: | Allison, T.M, Cochrane, F.C, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Examining the Role of Intersubunit Contacts in Catalysis by 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthase.
Biochemistry, 52, 2013
|
|
6HA1
 
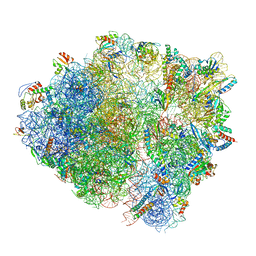 | | Cryo-EM structure of a 70S Bacillus subtilis ribosome translating the ErmD leader peptide in complex with telithromycin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Graf, M, Huter, P, Abdelshahid, M, Novacek, J, Wilson, D.N. | | Deposit date: | 2018-08-07 | | Release date: | 2018-08-29 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for antibiotic resistance mediated by theBacillus subtilisABCF ATPase VmlR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4JTF
 
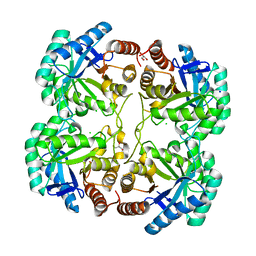 | | Crystal structure of F114R mutant of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Allison, T.M, Cochrane, F.C, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Examining the Role of Intersubunit Contacts in Catalysis by 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthase.
Biochemistry, 52, 2013
|
|
4WQ0
 
 | |
7AZY
 
 | |
3MOQ
 
 | |
4JDQ
 
 | |
4JTG
 
 | | Crystal structure of F114R/R117A mutant of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Allison, T.M, Cochrane, F.C, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Examining the Role of Intersubunit Contacts in Catalysis by 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthase.
Biochemistry, 52, 2013
|
|
4O0L
 
 | |
4H37
 
 | | Crystal structure of a voltage-gated K+ channel pore domain in a closed state in lipid membranes | | Descriptor: | Lmo2059 protein, POTASSIUM ION | | Authors: | Santos, J.S, Asmar-Rovira, G.A, Han, G.W, Liu, W, Syeda, R, Cherezov, V, Baker, K.A, Stevens, R.C, Montal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structure of a Voltage-gated K+ Channel Pore Module in a Closed State in Lipid Membranes.
J.Biol.Chem., 287, 2012
|
|
7PO4
 
 | | Assembly intermediate of human mitochondrial ribosome large subunit (largely unfolded rRNA with MALSU1, L0R8F8 and ACP) | | Descriptor: | 16SrRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Mechanism of mitoribosomal small subunit biogenesis and preinitiation.
Nature, 606, 2022
|
|
8DVW
 
 | | Structure of the Campylobacter concisus glycosyltransferase PglA R203Q | | Descriptor: | N, N'-diacetylbacillosaminyl-diphospho-undecaprenol alpha-1,3-N-acetylgalactosaminyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE | | Authors: | Vuksanovic, N, Clasman, J.R, Bernstein, H.M, Imperiali, B, Allen, K.N. | | Deposit date: | 2022-07-30 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Specificity determinants revealed by the structure of glycosyltransferase Campylobacter concisus PglA.
Protein Sci., 33, 2024
|
|
8DVZ
 
 | | Structure of the Campylobacter concisus glycosyltransferase PglA R282V variant | | Descriptor: | N, N'-diacetylbacillosaminyl-diphospho-undecaprenol alpha-1,3-N-acetylgalactosaminyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE | | Authors: | Vuksanovic, N, Clasman, J.R, Bernstein, H.M, Imperiali, B, Allen, K.N. | | Deposit date: | 2022-07-30 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Specificity determinants revealed by the structure of glycosyltransferase Campylobacter concisus PglA.
Protein Sci., 33, 2024
|
|
8DQD
 
 | | Structure of the Campylobacter concisus glycosyltransferase PglA | | Descriptor: | N, N'-diacetylbacillosaminyl-diphospho-undecaprenol alpha-1,3-N-acetylgalactosaminyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE | | Authors: | Vuksanovic, N, Clasman, J.R, Bernstein, H.M, Imperiali, B, Allen, K.N. | | Deposit date: | 2022-07-18 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Specificity determinants revealed by the structure of glycosyltransferase Campylobacter concisus PglA.
Protein Sci., 33, 2024
|
|
4GSL
 
 | | Crystal structure of an Atg7-Atg3 crosslinked complex | | Descriptor: | Autophagy-related protein 3, Ubiquitin-like modifier-activating enzyme ATG7, ZINC ION | | Authors: | Kaiser, S.E, Mao, K, Taherbhoy, A.M, Yu, S, Olszewski, J.L, Duda, D.M, Kurinov, I, Deng, A, Fenn, T.D, Klionsky, D.J, Schulman, B.A. | | Deposit date: | 2012-08-27 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7-Atg3 and Atg7-Atg10 structures.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AUP
 
 | | Crystal structure of Basic 7S globulin from soybean | | Descriptor: | Basic 7S globulin | | Authors: | Yoshizawa, T, Shimizu, T, Taichi, M, Nishiuchi, Y, Yamabe, M, Shichijo, N, Unzai, S, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-02-14 | | Release date: | 2011-04-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of basic 7S globulin, a xyloglucan-specific endo-beta-1,4-glucanase inhibitor protein-like protein from soybean lacking inhibitory activity against endo-beta-glucanase
Febs J., 278, 2011
|
|
3AW0
 
 | | Structure of SARS 3CL protease with peptidic aldehyde inhibitor | | Descriptor: | 3C-Like Proteinase, peptide ACE-SER-ALA-VAL-LEU-HIS-H | | Authors: | Akaji, K, Konno, H, Mitsui, H, Teruya, K, Hattori, Y, Ozaki, T, Kusunoki, M, Sanjho, A. | | Deposit date: | 2011-03-09 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design, Synthesis, and Evaluation of Peptide-Mimetic SARS 3CL Protease Inhibitors.
J.Med.Chem., 54, 2011
|
|
3PFK
 
 | |
3PDI
 
 | | Precursor bound NifEN | | Descriptor: | IRON/SULFUR CLUSTER, L-Cluster (Fe8S9), Nitrogenase MoFe cofactor biosynthesis protein NifE, ... | | Authors: | Kaiser, J.T, Hu, Y, Wiig, J.A, Rees, D.C, Ribbe, M.W. | | Deposit date: | 2010-10-22 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Precursor-Bound NifEN: A Nitrogenase FeMo Cofactor Maturase/Insertase.
Science, 331, 2011
|
|
4GSK
 
 | | Crystal structure of an Atg7-Atg10 crosslinked complex | | Descriptor: | Ubiquitin-like modifier-activating enzyme ATG7, Ubiquitin-like-conjugating enzyme ATG10, ZINC ION | | Authors: | Kaiser, S.E, Mao, K, Taherbhoy, A.M, Yu, S, Olszewski, J.L, Duda, D.M, Kurinov, I, Deng, A, Fenn, T.D, Klionsky, D.J, Schulman, B.A. | | Deposit date: | 2012-08-27 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7-Atg3 and Atg7-Atg10 structures.
Nat.Struct.Mol.Biol., 19, 2012
|
|
8OSE
 
 | | C. perfringens chitinase CP4_3455 in complex with inhibitor bisdionin C | | Descriptor: | 1,1'-PROPANE-1,3-DIYLBIS(3,7-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE), 1,2-ETHANEDIOL, Chitodextrinase, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-04-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | C. perfringens chitinase CP4_3455 in complex with inhibitor bisdionin C
To Be Published
|
|
