7EWD
 
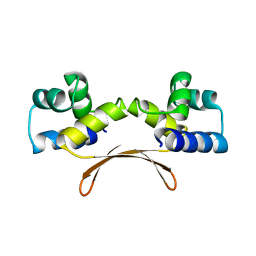 | | Mycobacterium tuberculosis HigA2 (Form II) | | Descriptor: | Putative antitoxin HigA2 | | Authors: | Kim, H.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chasing the structural diversity of the transcription regulator Mycobacterium tuberculosis HigA2.
Iucrj, 8, 2021
|
|
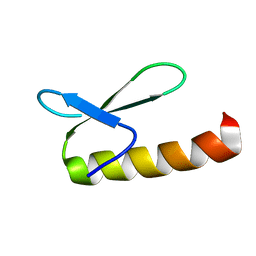
1KJK
 
 | |
4IWG
 
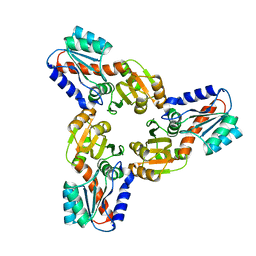 | | Crystal Structure of the Conserved Hypothetical Protein MJ0927 from Methanocaldococcus jannaschii (in C2221 form) | | Descriptor: | UPF0135 protein MJ0927 | | Authors: | Kuan, S.M, Chen, S.C, Yang, C.S, Chen, Y.R, Liu, Y.H, Chen, Y. | | Deposit date: | 2013-01-23 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Crystal structure of a conserved hypothetical protein MJ0927 from Methanocaldococcus jannaschii reveals a novel quaternary assembly in the Nif3 family.
Biomed Res Int, 2014, 2014
|
|
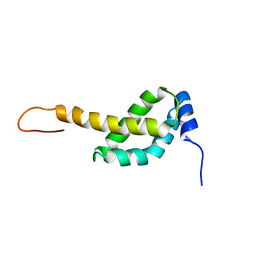
2MAO
 
 | |
2H3C
 
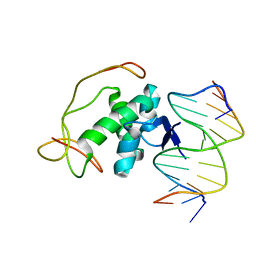 | | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*TP*AP*TP*AP*CP*CP*CP*G)-3', 5'-D(P*TP*CP*GP*GP*GP*TP*AP*TP*AP*CP*AP*TP*A)-3', CcdA | | Authors: | Madl, T, Van Melderen, L, Respondek, M, Oberer, M, Keller, W, Zangger, K. | | Deposit date: | 2006-05-22 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Nucleic Acid and Toxin Recognition of the Bacterial Antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2KZ8
 
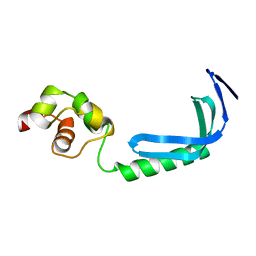 | | Solution NMR structure of MqsA, a protein from E. coli, containing a Zinc finger, N-terminal and a Helix Turn-Helix C-terminal domain | | Descriptor: | Uncharacterized HTH-type transcriptional regulator ygiT | | Authors: | Papadopoulos, E, Vlamis-Gardikas, A, Graslund, A, Billeter, M, Holmgren, A, Collet, J. | | Deposit date: | 2010-06-14 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and biophysical properties of MqsA, a Zn-containing antitoxin from Escherichia coli
Biochim.Biophys.Acta, 2012
|
|
7OGT
 
 | | Folded elbow of cohesin | | Descriptor: | Structural maintenance of chromosomes protein 1, Structural maintenance of chromosomes protein 3 | | Authors: | Lee, B.-G, Gonzalez Llamazares, A, Collier, J, Patele, N.J, Nasmyth, K.A, Lowe, J. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Folding of cohesin's coiled coil is important for Scc2/4-induced association with chromosomes.
Elife, 10, 2021
|
|
6QKQ
 
 | |
6QKP
 
 | |
6TC0
 
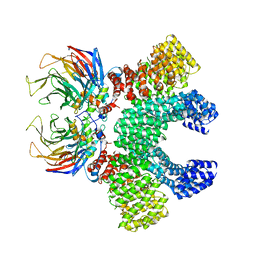 | | Crystal structure of MMS19-CIAO1-CIAO2B CIA targeting complex | | Descriptor: | MIP18 family protein galla-2, MMS19 nucleotide excision repair protein homolog, Probable cytosolic iron-sulfur protein assembly protein Ciao1 | | Authors: | Kassube, S.A, Thoma, N.H. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insights into Fe-S protein biogenesis by the CIA targeting complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8SP7
 
 | |
8SP5
 
 | |
7QAA
 
 | | Crystal structure of RARalpha/RXRalpha ligand binding domain heterodimer in complex with BMS614 and oleic acid | | Descriptor: | 4-[(4,4-DIMETHYL-1,2,3,4-TETRAHYDRO-[1,2']BINAPTHALENYL-7-CARBONYL)-AMINO]-BENZOIC ACID, Isoform Alpha-1-deltaBC of Retinoic acid receptor alpha, OLEIC ACID, ... | | Authors: | le Maire, A, Vivat, V, Guee, L, Blanc, P, Malosse, C, Chamot-Rooke, J, Germain, P, Bourguet, w. | | Deposit date: | 2021-11-16 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Design and in vitro characterization of RXR variants as tools to investigate the biological role of endogenous rexinoids.
J.Mol.Endocrinol., 69, 2022
|
|
4ZWQ
 
 | |
8ISY
 
 | | Cryo-EM structure of free-state Crt-SPARTA | | Descriptor: | Piwi domain-containing protein, TIR domain-containing protein | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
4ZWR
 
 | |
4ZWS
 
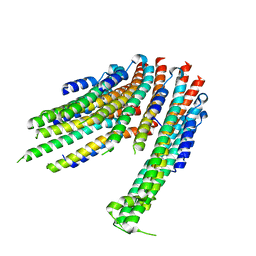 | |
8SZP
 
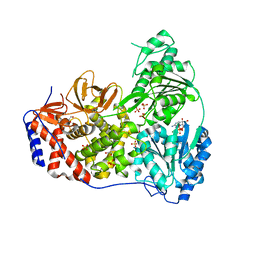 | | Human DHX9 bound to ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase A, ... | | Authors: | Lee, Y.-T, Sickmier, E.A, Grigoriu, S, Boriack-Sjodin, P.A. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structures of the DExH-box RNA helicase DHX9.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6OD7
 
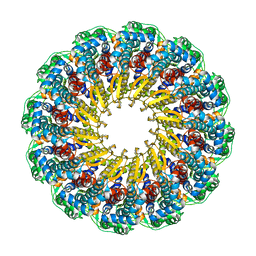 | | Herpes simplex virus type 1 (HSV-1) pUL6 portal protein, dodecameric complex | | Descriptor: | Portal protein | | Authors: | Liu, Y.T, Jih, J, Dai, X.H, Bi, G.Q, Zhou, Z.H. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structures of herpes simplex virus type 1 portal vertex and packaged genome.
Nature, 570, 2019
|
|
5MEZ
 
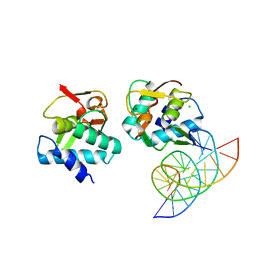 | | Crystal structure of Smad4-MH1 bound to the GGCT site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*GP*CP*AP*GP*GP*CP*TP*AP*GP*CP*CP*TP*GP*CP*A)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5YHY
 
 | | Structure of Lactococcus lactis ZitR, C30S mutant | | Descriptor: | ZINC ION, Zinc transport transcriptional regulator | | Authors: | Song, Y, Liu, H, Zhu, R, Yi, C, Chen, P. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation.
Proc.Natl.Acad.Sci.USA, 114, 2017
|
|
5AIQ
 
 | | Crystal structure of ligand-free NadR | | Descriptor: | TRANSCRIPTIONAL REGULATOR, MARR FAMILY | | Authors: | Liguori, A, Malito, E, Bottomley, M.J. | | Deposit date: | 2015-02-16 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.716 Å) | | Cite: | Molecular Basis of Ligand-Dependent Regulation of Nadr, the Transcriptional Repressor of Meningococcal Virulence Factor Nada.
Plos Pathog., 12, 2016
|
|
5YHZ
 
 | | Structure of Lactococcus lactis ZitR, E41A mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ZINC ION, ... | | Authors: | Song, Y, Liu, H, Zhu, R, Yi, C, Chen, P. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation.
Proc.Natl.Acad.Sci.USA, 114, 2017
|
|
5AIP
 
 | | Crystal structure of NadR in complex with 4-hydroxyphenylacetate | | Descriptor: | 4-HYDROXYPHENYLACETATE, TRANSCRIPTIONAL REGULATOR, MARR FAMILY | | Authors: | Liguori, A, Malito, E, Bottomley, M.J. | | Deposit date: | 2015-02-16 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis of Ligand-Dependent Regulation of Nadr, the Transcriptional Repressor of Meningococcal Virulence Factor Nada.
Plos Pathog., 12, 2016
|
|
5MEY
 
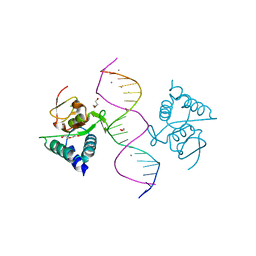 | | Crystal structure of Smad4-MH1 bound to the GGCGC site. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
