1D89
 
 | |
6X1V
 
 | | Structure of pHis Fab (SC44-8) in complex with pHis mimetic peptide | | Descriptor: | 1,2-ETHANEDIOL, ACLYana-3-pTza peptide, SC44-8 Heavy chain, ... | | Authors: | Kalagiri, R, Stanfield, R, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5F0E
 
 | | Murine endoplasmic reticulum alpha-glucosidase II | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2015-11-27 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1D9M
 
 | |
1CE9
 
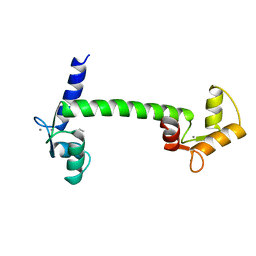
 | | HELIX CAPPING IN THE GCN4 LEUCINE ZIPPER | | Descriptor: | PROTEIN (GCN4-PMSE) | | Authors: | Lu, M, Shu, W, Ji, H, Spek, E, Wang, L.-Y, Kallenbach, N.R. | | Deposit date: | 1999-03-18 | | Release date: | 1999-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Helix capping in the GCN4 leucine zipper.
J.Mol.Biol., 288, 1999
|
|
1CLL
 
 | |
6WK2
 
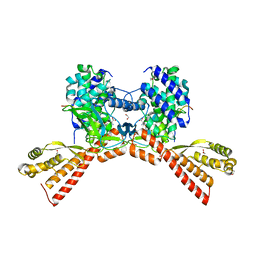 | | SETD3 mutant (N255V) in Complex with an Actin Peptide with His73 Replaced with Methionine | | Descriptor: | 1,2-ETHANEDIOL, Actin, cytoplasmic 2, ... | | Authors: | Dai, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Characterization of SETD3 methyltransferase-mediated protein methionine methylation.
J.Biol.Chem., 295, 2020
|
|
5F11
 
 | |
6WVV
 
 | | Plasmodium vivax M17 leucyl aminopeptidase | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, M17 leucyl aminopeptidase, ... | | Authors: | Malcolm, T.R, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Active site metals mediate an oligomeric equilibrium in Plasmodium M17 aminopeptidases.
J.Biol.Chem., 296, 2020
|
|
2INE
 
 | |
6X1T
 
 | | Structure of pHis Fab (SC50-3) in complex with pHis mimetic peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5EVY
 
 | | Salicylate hydroxylase substrate complex | | Descriptor: | 2-HYDROXYBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Salicylate hydroxylase | | Authors: | Morimoto, Y, Uemura, T. | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The catalytic mechanism of decarboxylative hydroxylation of salicylate hydroxylase revealed by crystal structure analysis at 2.5 angstrom resolution
Biochem.Biophys.Res.Commun., 469, 2016
|
|
3I8W
 
 | |
6X3C
 
 | | Crystal structure of streptogramin A acetyltransferase VatA from Staphylococcus aureus in complex with streptogramin analog F1037 (47) | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chaires, H.A, Fraser, J.S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
6WML
 
 | | Human TLR8 bound to the potent agonist, GS-9688 (Selgantolimod) | | Descriptor: | (2R)-2-[(2-amino-7-fluoropyrido[3,2-d]pyrimidin-4-yl)amino]-2-methylhexan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Appleby, T.C, Perry, J.K, Mish, M, Villasenor, A.G, Mackman, R.L. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of GS-9688 (Selgantolimod) as a Potent and Selective Oral Toll-Like Receptor 8 Agonist for the Treatment of Chronic Hepatitis B.
J.Med.Chem., 63, 2020
|
|
1CLF
 
 | | CLOSTRIDIUM PASTEURIANUM FERREDOXIN | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Bertini, I, Donaire, A, Feinberg, B.A, Luchinat, C, Piccioli, M, Yuan, H. | | Deposit date: | 1995-06-21 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized 2[4Fe-4S] ferredoxin from Clostridium pasteurianum.
Eur.J.Biochem., 232, 1995
|
|
1U9Q
 
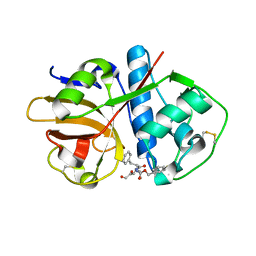 | | Crystal structure of cruzain bound to an alpha-ketoester | | Descriptor: | [1-(1-METHYL-4,5-DIOXO-PENT-2-ENYLCARBAMOYL)-2-PHENYL-ETHYL]-CARBAMIC ACID BENZYL ESTER, cruzipain | | Authors: | Lange, M, Weston, S.G, Cheng, H, Culliane, M, Fiorey, M.M, Grisostomi, C, Hardy, L.W, Hartstough, D.S, Pallai, P.V, Tilton, R.F, Baldino, C.M, Brinen, L.S, Engel, J.C, Choe, Y, Price, M.S, Craik, C.S. | | Deposit date: | 2004-08-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of alpha-keto-based inhibitors of cruzain, a cysteine protease implicated in Chagas disease
Bioorg.Med.Chem., 13, 2005
|
|
6WUF
 
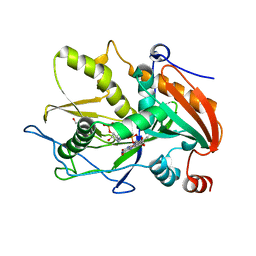 | | Crystal structure of mouse DXO in complex with 3'-FADP | | Descriptor: | 1,2-ETHANEDIOL, Decapping and exoribonuclease protein, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Doamekpor, S.K, Tong, L. | | Deposit date: | 2020-05-04 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DXO/Rai1 enzymes remove 5'-end FAD and dephospho-CoA caps on RNAs.
Nucleic Acids Res., 48, 2020
|
|
7T71
 
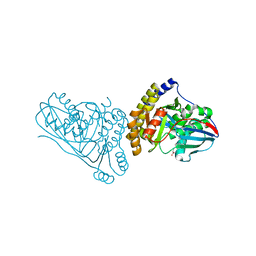 | | Crystal Structure of Mevalonate 3,5-Bisphosphate Decarboxylase from Picrophilus Torridus | | Descriptor: | Mevalonate 3,5-bisphosphate decarboxylase, OLEIC ACID | | Authors: | Vinokur, J.M, Sawaya, M.R, Cascio, D, Collazo, M, Bowie, J.U. | | Deposit date: | 2021-12-14 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of mevalonate 3,5-bisphosphate decarboxylase reveals insight into the evolution of decarboxylases in the mevalonate metabolic pathways.
J.Biol.Chem., 298, 2022
|
|
5EYN
 
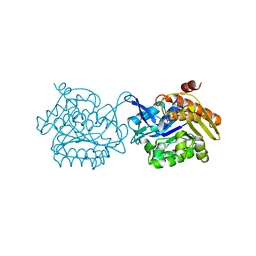 | | Crystal structure of Fructokinase from Vibrio cholerae O395 in fructose, ADP, Beryllium trifluoride and calcium ion bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Paul, R, Nath, S, Sen, U. | | Deposit date: | 2015-11-25 | | Release date: | 2016-11-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.289 Å) | | Cite: | Crystal structure of Fructokinase from Vibrio cholerae O395 in apo form
To Be Published
|
|
7TE8
 
 | | CA14-CBD-DB21 ternary complex | | Descriptor: | CA14, DB21, cannabidiol | | Authors: | Cao, S, Zheng, N. | | Deposit date: | 2022-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Defining molecular glues with a dual-nanobody cannabidiol sensor.
Nat Commun, 13, 2022
|
|
6WXX
 
 | | crystal structure of cA4-activated Card1 | | Descriptor: | Card1, MANGANESE (II) ION, cA4 | | Authors: | Rostol, J, Xie, W, Patel, D.J, Marraffini, L. | | Deposit date: | 2020-05-12 | | Release date: | 2020-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Card1 nuclease provides defence during type III CRISPR immunity.
Nature, 590, 2021
|
|
5F0Z
 
 | |
6X3J
 
 | | Crystal structure of streptogramin A acetyltransferase VatA from Staphylococcus aureus in complex with streptogramin analog F0224 (46) | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-4,12-dimethyl-1,7,22-trioxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl isoquinolin-3-ylcarbamate, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chaires, H.A, Fraser, J.S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
6HWU
 
 | | Crystal structure of p38alpha in complex with a photoswitchable 2-Azothiazol-based Inhibitor (compound 2) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-[(~{E})-phenyldiazenyl]-1,3-thiazol-5-yl]pyridin-2-yl]propanamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Mueller, M.P, Rauh, D. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
