1YLD
 
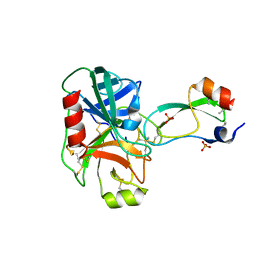 | | Trypsin/BPTI complex mutant | | Descriptor: | CALCIUM ION, Pancreatic trypsin inhibitor, SULFATE ION, ... | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-19 | | Release date: | 2006-04-25 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Partially folded bovine pancreatic trypsin inhibitor analogues attain fully native structures when co-crystallized with S195A rat trypsin
J.Mol.Biol., 375, 2008
|
|
5MPD
 
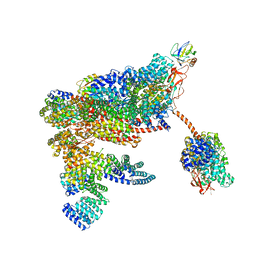 | | 26S proteasome in presence of ATP (s1) | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit RPN1, 26S proteasome regulatory subunit RPN10, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4H37
 
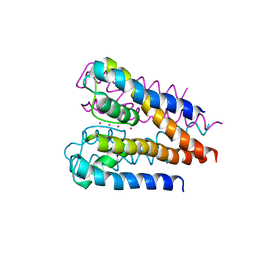 | | Crystal structure of a voltage-gated K+ channel pore domain in a closed state in lipid membranes | | Descriptor: | Lmo2059 protein, POTASSIUM ION | | Authors: | Santos, J.S, Asmar-Rovira, G.A, Han, G.W, Liu, W, Syeda, R, Cherezov, V, Baker, K.A, Stevens, R.C, Montal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structure of a Voltage-gated K+ Channel Pore Module in a Closed State in Lipid Membranes.
J.Biol.Chem., 287, 2012
|
|
7AQO
 
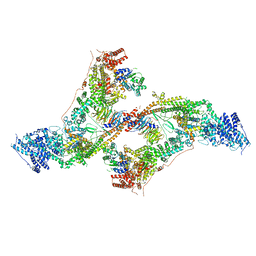 | | yeast THO-Sub2 complex dimer | | Descriptor: | BJ4_G0025130.mRNA.1.CDS.1, EM14S01-3B_G0007820.mRNA.1.CDS.1, TEX1 isoform 1, ... | | Authors: | Schuller, S.K, Schuller, J.M, Prabu, R.J, Baumgartner, M, Bonneau, F, basquin, J, Conti, E. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex.
Elife, 9, 2020
|
|
1YLC
 
 | | Trypsin/BPTI complex mutant | | Descriptor: | CALCIUM ION, Pancreatic trypsin inhibitor, SULFATE ION, ... | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-19 | | Release date: | 2006-04-25 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Partially folded bovine pancreatic trypsin inhibitor analogues attain fully native structures when co-crystallized with S195A rat trypsin
J.Mol.Biol., 375, 2008
|
|
1YLH
 
 | | Crystal Structure of Phosphoenolpyruvate Carboxykinase from Actinobaccilus succinogenes in Complex with Manganese and Pyruvate | | Descriptor: | (2S,3S)-2,3-DIHYDROXY-4-SULFANYLBUTANE-1-SULFONATE, BETA-MERCAPTOETHANOL, FORMIC ACID, ... | | Authors: | Leduc, Y.A, Prasad, L, Laivenieks, M, Zeikus, J.G, Delbaere, L.T. | | Deposit date: | 2005-01-19 | | Release date: | 2005-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of PEP carboxykinase from the succinate-producing Actinobacillus succinogenes: a new conserved active-site motif.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3OKH
 
 | |
5MD2
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=5, twist=-6 | | Descriptor: | Capsid protein p24 C-terminal domain, Capsid protein p24 N-terminal domain | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MDC
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=17, twist=-12 | | Descriptor: | Gag protein | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
1YO8
 
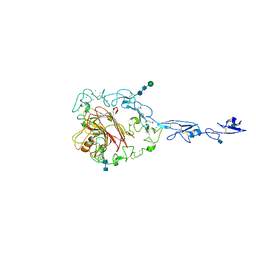 | | Structure of the C-terminal domain of human thrombospondin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Carlson, C.B, Bernstein, D.A, Annis, D.S, Misenheimer, T.M, Hannah, B.A, Mosher, D.F, Keck, J.L. | | Deposit date: | 2005-01-26 | | Release date: | 2005-09-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the calcium-rich signature domain of human thrombospondin-2
Nat.Struct.Mol.Biol., 12, 2005
|
|
3ME6
 
 | | Crystal structure of cytochrome 2B4 in complex with the anti-platelet drug clopidogrel | | Descriptor: | Clopidogrel, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gay, S.C, Roberts, A.G, Maekawa, K, Talakad, J.C, Hong, W.X, Zhang, Q, Stout, C.D, Halpert, J.R. | | Deposit date: | 2010-03-31 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of cytochrome P450 2B4 complexed with the antiplatelet drugs ticlopidine and clopidogrel.
Biochemistry, 49, 2010
|
|
3MG4
 
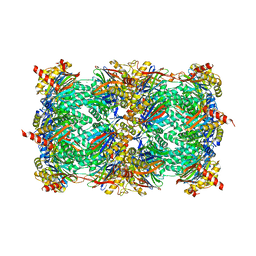 | | Structure of yeast 20S proteasome with Compound 1 | | Descriptor: | (2S)-2-amino-N-[(1S)-1-({(1S)-1-[(4-methylbenzyl)carbamoyl]-3-phenylpropyl}carbamoyl)-3-phenylpropyl]-4-phenylbutanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-04-05 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
1YN4
 
 | | Crystal Structures of EAP Domains from Staphylococcus aureus Reveal an Unexpected Homology to Bacterial Superantigens | | Descriptor: | EapH1, ZINC ION | | Authors: | Geisbrecht, B.V, Hamaoka, B.Y, Perman, B, Zemla, A, Leahy, D.J. | | Deposit date: | 2005-01-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structures of EAP Domains from Staphylococcus aureus Reveal an Unexpected Homology to Bacterial Superantigens.
J.Biol.Chem., 280, 2005
|
|
1YNB
 
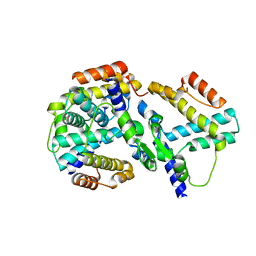 | | crystal structure of genomics APC5600 | | Descriptor: | hypothetical protein AF1432 | | Authors: | Dong, A, Skarina, T, Savchenko, A, Pai, E.F, Joachimiak, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-01-24 | | Release date: | 2005-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of genomics AF1432 by Sulfur SAD methods
To be Published
|
|
1YNN
 
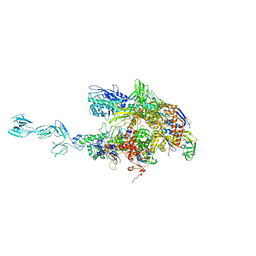 | | Taq RNA polymerase-rifampicin complex | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Campbell, E.A, Pavlova, O, Zenkin, N, Leon, F, Irschik, H, Jansen, R, Severinov, K, Darst, S.A. | | Deposit date: | 2005-01-24 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural, functional, and genetic analysis of sorangicin inhibition of bacterial RNA polymerase
Embo J., 24, 2005
|
|
1YQP
 
 | | T268N mutant cytochrome domain of flavocytochrome P450 BM3 | | Descriptor: | Bifunctional P-450:NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Clark, J.P, Miles, C.S, Mowat, C.G, Walkinshaw, M.D, Reid, G.A, Daff, S.N, Chapman, S.K. | | Deposit date: | 2005-02-02 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The role of Thr268 and Phe393 in cytochrome P450 BM3.
J.Inorg.Biochem., 100, 2006
|
|
4IK3
 
 | | High resolution structure of GCaMP3 at pH 8.5 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
3M5M
 
 | |
3MM0
 
 | | Crystal structure of chimeric avidin | | Descriptor: | Avidin, Avidin-related protein 4/5 | | Authors: | Livnah, O, Eisenberg-Domovich, Y, Maatta, J.A.E, Kulomaa, M.S, Hytonen, V.P, Nordlund, H.R. | | Deposit date: | 2010-04-19 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chimeric avidin shows stability against harsh chemical conditions-biochemical analysis and 3D structure.
Biotechnol.Bioeng., 108, 2011
|
|
3HUG
 
 | |
4IFD
 
 | |
3HTO
 
 | | the hemagglutinin structure of an avian H1N1 influenza A virus | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Wang, G, Li, A, Zhang, Q, Wu, C, Zhang, R, Cai, Q, Song, W, Yuen, K.-Y. | | Deposit date: | 2009-06-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The hemagglutinin structure of an avian H1N1 influenza A virus
Virology, 392, 2009
|
|
3HTY
 
 | |
4INR
 
 | | Yeast 20S proteasome in complex with the vinyl sulfone LU102 | | Descriptor: | N3Phe-Leu-Leu-Phe(4-NH2CH2)-methyl vinyl sulfone, bound form, Proteasome component C1, ... | | Authors: | Geurink, P.P, van der Linden, W.A, Mirabella, A.C, Gallastegui, N, de Bruin, G, Blom, A.E.M, Voges, M.J, Mock, E.D, Florea, B.I, van der Marel, G.A, Driessen, C, van der Stelt, M, Groll, M, Overkleeft, H.S, Kisselev, A.F. | | Deposit date: | 2013-01-06 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Incorporation of Non-natural Amino Acids Improves Cell Permeability and Potency of Specific Inhibitors of Proteasome Trypsin-like Sites.
J.Med.Chem., 56, 2013
|
|
4INT
 
 | | Yeast 20S proteasome in complex with the vinyl sulfone LU122 | | Descriptor: | HMB-Val-Ser-Phe(4-NH2CH2)-methyl vinyl sulfone, bound form, Proteasome component C1, ... | | Authors: | Geurink, P.P, van der Linden, W.A, Mirabella, A.C, Gallastegui, N, de Bruin, G, Blom, A.E.M, Voges, M.J, Mock, E.D, Florea, B.I, van der Marel, G.A, Driessen, C, van der Stelt, M, Groll, M, Overkleeft, H.S, Kisselev, A.F. | | Deposit date: | 2013-01-06 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Incorporation of Non-natural Amino Acids Improves Cell Permeability and Potency of Specific Inhibitors of Proteasome Trypsin-like Sites.
J.Med.Chem., 56, 2013
|
|
