2OOV
 
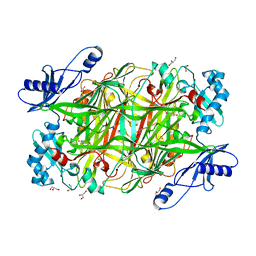 | |
2OPS
 
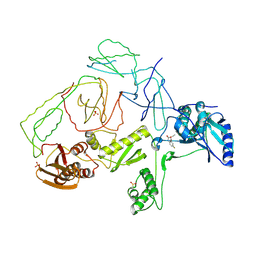 | | Crystal Structure of Y188C Mutant HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Short, S.A, Chan, J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
4CWZ
 
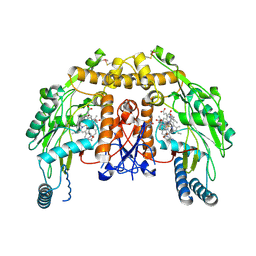 | | Structure of bovine endothelial nitric oxide synthase Y477A mutant heme domain in complex with 4-METHYL-6-(((3R,4R)-4-((5-(4- METHYLPYRIDIN-2-YL)PENTYL)OXY)PYRROLIDIN-3-YL)METHYL)PYRIDIN-2-AMINE | | Descriptor: | 4-methyl-6-{[(3R,4R)-4-{[5-(4-methylpyridin-2-yl)pentyl]oxy}pyrrolidin-3-yl]methyl}pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-04-03 | | Release date: | 2014-08-13 | | Last modified: | 2014-09-03 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Mobility of a Conserved Tyrosine Residue Controls Isoform-Dependent Enzyme-Inhibitor Interactions in Nitric Oxide Synthases.
Biochemistry, 53, 2014
|
|
2OQD
 
 | | Crystal Structure of BthTX-II | | Descriptor: | Phospholipase A2 | | Authors: | Correa, L.C, Marchi-Salvador, D.P, Cintra, A.C.O, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2007-01-31 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of a myotoxic Asp49-phospholipase A(2) with low catalytic activity: Insights into Ca(2+)-independent catalytic mechanism.
Biochim.Biophys.Acta, 1784, 2008
|
|
2ORQ
 
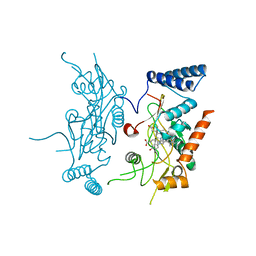 | | Murine Inducible Nitric Oxide Synthase Oxygenase Domain (DELTA 114) 4-(imidazol-1-yl)phenol and piperonylamine Complex | | Descriptor: | 1-(1,3-BENZODIOXOL-5-YL)METHANAMINE, 4-(1H-IMIDAZOL-1-YL)PHENOL, Nitric oxide synthase, ... | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2007-02-04 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Activity of 2-Imidazol-1-ylpyrimidine Derived Inducible Nitric Oxide Synthase Dimerization Inhibitors
J.Med.Chem., 50, 2007
|
|
3CNE
 
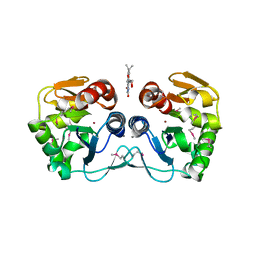 | | Crystal structure of the putative protease I from Bacteroides thetaiotaomicron | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, Putative protease I, ... | | Authors: | Zhang, R, Volkart, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the putative protease I from Bacteroides thetaiotaomicron.
To be Published
|
|
3CO7
 
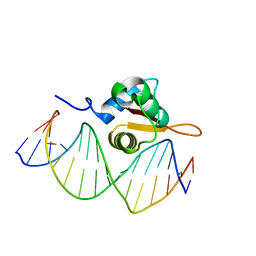 | | Crystal Structure of FoxO1 DBD Bound to DBE2 DNA | | Descriptor: | DNA (5'-D(*DCP*DAP*DAP*DAP*DAP*DTP*DGP*DTP*DAP*DAP*DAP*DCP*DAP*DAP*DGP*DA)-3'), DNA (5'-D(*DTP*DCP*DTP*DTP*DGP*DTP*DTP*DTP*DAP*DCP*DAP*DTP*DTP*DTP*DTP*DG)-3'), Forkhead box protein O1 | | Authors: | Brent, M.M, Anand, R, Marmorstein, R. | | Deposit date: | 2008-03-27 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Basis for DNA Recognition by FoxO1 and Its Regulation by Posttranslational Modification.
Structure, 16, 2008
|
|
4D2E
 
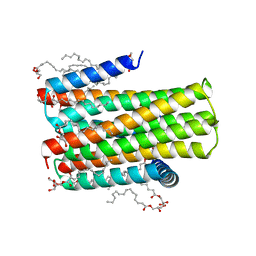 | | Crystal structure of an integral membrane kinase - v2.3 | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, CITRATE ANION, ... | | Authors: | Li, D, Boland, C, Caffrey, M. | | Deposit date: | 2014-05-09 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Cell-Free Expression and in Meso Crystallisation of an Integral Membrane Kinase for Structure Determination.
Cell.Mol.Life Sci., 71, 2014
|
|
2O25
 
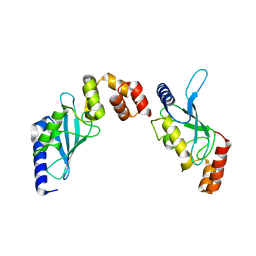 | | Ubiquitin-Conjugating Enzyme E2-25 kDa Complexed With SUMO-1-Conjugating Enzyme UBC9 | | Descriptor: | SUMO-1-conjugating enzyme UBC9, Ubiquitin-conjugating enzyme E2-25 kDa | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-29 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel and Unexpected Complex Between the SUMO-1-Conjugating Enzyme UBC9 and the Ubiquitin-Conjugating Enzyme E2-25 kDa
To be Published
|
|
4D35
 
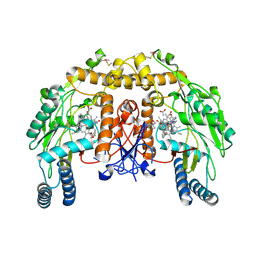 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with N-2-(2-(1H-imidazol-1-yl)pyrimidin-4-yl)ethyl-3-(3- fluorophenyl)propan-1-amine | | Descriptor: | 3-(3-fluorophenyl)-N-{2-[2-(1H-imidazol-1-yl)pyrimidin-4-yl]ethyl}propan-1-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-10-20 | | Release date: | 2014-12-24 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Novel 2,4-Disubstituted Pyrimidines as Potent, Selective, and Cell-Permeable Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
2NT6
 
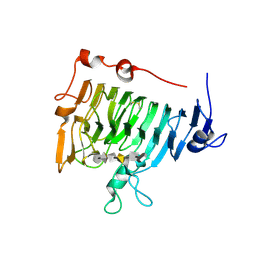 | | Crystal structure of pectin methylesterase D178A mutant in complex with hexasaccharide III | | Descriptor: | Pectinesterase A, methyl alpha-D-galactopyranuronate-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-methyl alpha-D-galactopyranuronate | | Authors: | Fries, M, Brocklehurst, K, Shevchik, V.E, Pickersgill, R.W. | | Deposit date: | 2006-11-07 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of the activity of the phytopathogen pectin methylesterase.
Embo J., 26, 2007
|
|
6NP0
 
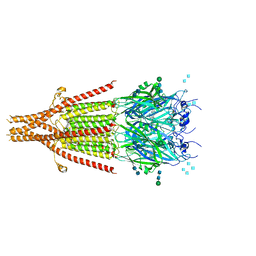 | | Cryo-EM structure of 5HT3A receptor in presence of granisetron | | Descriptor: | 1-methyl-N-[(1R,5S)-9-methyl-9-azabicyclo[3.3.1]nonan-3-yl]indazole-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Basak, S, Chakrapani, S. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Molecular mechanism of setron-mediated inhibition of full-length 5-HT3Areceptor.
Nat Commun, 10, 2019
|
|
2O2I
 
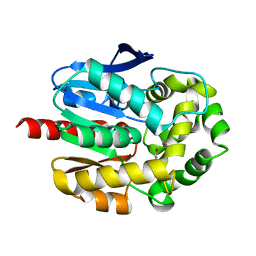 | | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,3-propandiol | | Descriptor: | 1,3-PROPANDIOL, BROMIDE ION, Haloalkane dehalogenase 3 | | Authors: | Mazumdar, P.A, Hulecki, J, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-11-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,3-propandiol
To be Published
|
|
4CTR
 
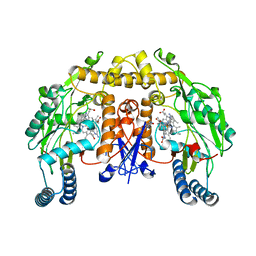 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 2-(6-Amino-4-methylpyridin-2-yl)-1-(3-(2-(6-amino-4- methylpyridin-2-yl)ethyl )phenyl)ethan-1-ol | | Descriptor: | (1S)-2-(6-amino-4-methylpyridin-2-yl)-1-{3-[2-(6-amino-4-methylpyridin-2-yl)ethyl]phenyl}ethanol, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-03-15 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nitric Oxide Synthase Inhibitors that Interact with Both a Heme Propionate and Tetrahydrobiopterin Show High Isoform Selectivity.
J.Med.Chem., 57, 2014
|
|
2NTW
 
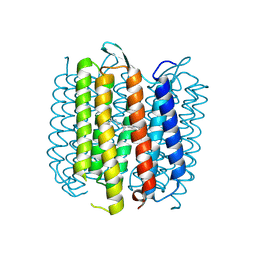 | |
3CJB
 
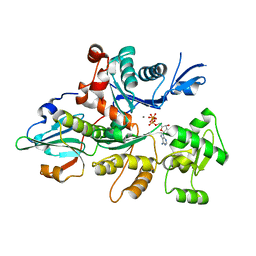 | | Actin dimer cross-linked by V. cholerae MARTX toxin and complexed with Gelsolin-segment 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Sawaya, M.R, Kudryashov, D.S, Pashkov, I, Reisler, E, Yeates, T.O. | | Deposit date: | 2008-03-12 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Connecting actin monomers by iso-peptide bond is a toxicity mechanism of the Vibrio cholerae MARTX toxin.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6T9P
 
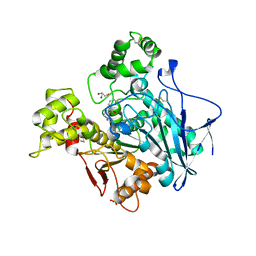 | | Human Butyrylcholinesterase in complex with 2-(N-hydroxyimino)-N-[(1R)-3-{4-[(2-methyl-1H-imidazol-1-yl)methyl]-1H-1,2,3-triazol-1-yl}-1- phenylpropyl]acetamide | | Descriptor: | (R,E)-2-(hydroxyimino)-N-(3-(4-((2-methyl-1H-imidazol-1-yl)methyl)-1H-1,2,3-triazol-1-yl)-1-phenylpropyl)acetamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[1-deoxy-alpha-D-tagatopyranose-(2-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Sinko, G, Marakovic, N, Knezevic, A. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enantioseparation, in vitro testing, and structural characterization of triple-binding reactivators of organophosphate-inhibited cholinesterases.
Biochem.J., 477, 2020
|
|
6NSE
 
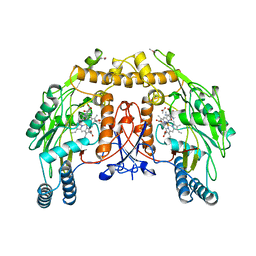 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, H4B-FREE, CANAVANINE COMPLEX | | Descriptor: | ACETATE ION, CACODYLIC ACID, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-01-13 | | Release date: | 2002-05-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures of the Heme Domain of Bovine Endothelial Nitric Oxide Synthase Complexed with Arginine Analogues
To be Published
|
|
2NVE
 
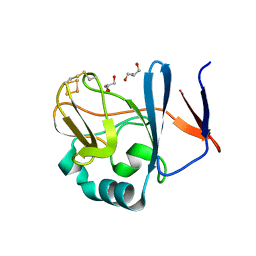 | | Soluble domain of Rieske Iron Sulfur Protein | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | Authors: | Kolling, D.K, Brunzelle, J.S, Lhee, S, Crofts, A.R, Nair, S.K. | | Deposit date: | 2006-11-12 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
4CXJ
 
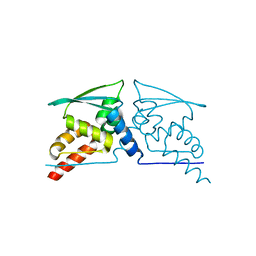 | | BTB domain of KEAP1 C151W mutant | | Descriptor: | KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Cleasby, A, Yon, J, Day, P.J, Richardson, C, Tickle, I.J, Williams, P.A, Callahan, J.F, Carr, R, Concha, N, Kerns, J.K, Qi, H, Sweitzer, T, Ward, P, Davies, T.G. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Btb Domain of Keap1 and its Interaction with the Triterpenoid Antagonist Cddo.
Plos One, 9, 2014
|
|
2NVG
 
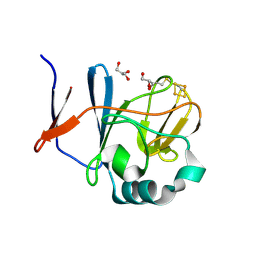 | | Soluble domain of Rieske Iron Sulfur protein. | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | Authors: | Kolling, D, Brunzelle, J, Lhee, S, Crofts, A.R, Nair, S.K. | | Deposit date: | 2006-11-12 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
2O5E
 
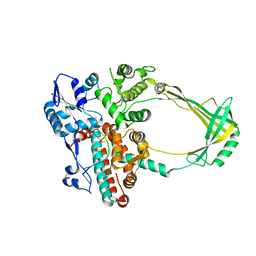 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glucose pH 7.0 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', CHLORIDE ION, DNA topoisomerase 3, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
6NTL
 
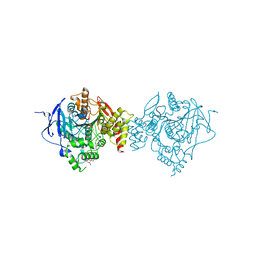 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by A-234 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Bester, S.M, Guelta, M.A, Height, J.J, Pegan, S.D. | | Deposit date: | 2019-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights into inhibition of human acetylcholinesterase by Novichok, A-series Nerve Agents
To Be Published
|
|
2NXT
 
 | |
3CVW
 
 | | Drosophila melanogaster (6-4) photolyase H365N mutant bound to ds DNA with a T-T (6-4) photolesion and cofactor F0 | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of a cofactor f0
accelerated (6-4) photolyase from the fruit fly
To be Published
|
|
