7Q65
 
 | | Cryo-em structure of the Nup98 fibril polymorph 2 | | Descriptor: | Nuclear pore complex protein Nup98 | | Authors: | Ibanez de Opakua, A, Geraets, J.A, Frieg, B, Dienemann, C, Savastano, A, Rankovic, M, Cima-Omori, M.-S, Schroeder, G.F, Zweckstetter, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular interactions of FG nucleoporin repeats at high resolution.
Nat.Chem., 14, 2022
|
|
7Q67
 
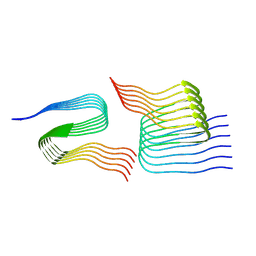 | | Cryo-em structure of the Nup98 fibril polymorph 4 | | Descriptor: | Nuclear pore complex protein Nup98 | | Authors: | Ibanez de Opakua, A, Geraets, J.A, Frieg, B, Dienemann, C, Savastano, A, Rankovic, M, Cima-Omori, M.-S, Schroeder, G.F, Zweckstetter, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Molecular interactions of FG nucleoporin repeats at high resolution.
Nat.Chem., 14, 2022
|
|
3R34
 
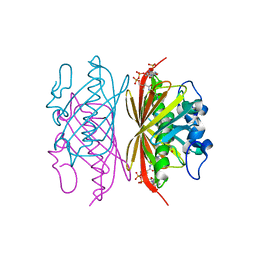 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase mutant E73D complexed with CoA | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxybenzoyl-CoA thioesterase, COENZYME A | | Authors: | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
3R3F
 
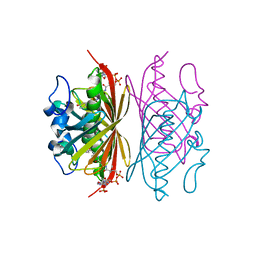 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase mutant T77A complexed with 4-hydroxyphenacyl CoA | | Descriptor: | 4-HYDROXYPHENACYL COENZYME A, 4-hydroxybenzoyl-CoA thioesterase | | Authors: | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
7Q66
 
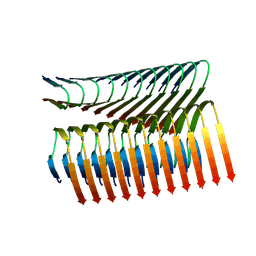 | | Cryo-em structure of the Nup98 fibril polymorph 3 | | Descriptor: | Nuclear pore complex protein Nup98 | | Authors: | Ibanez de Opakua, A, Geraets, J.A, Frieg, B, Dienemann, C, Savastano, A, Rankovic, M, Cima-Omori, M.-S, Schroeder, G.F, Zweckstetter, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Molecular interactions of FG nucleoporin repeats at high resolution.
Nat.Chem., 14, 2022
|
|
7PZO
 
 | | mite allergen Der p 3 from Dermatophagoides pteronyssinus | | Descriptor: | SULFATE ION, mite allergen Der p 3 | | Authors: | Timofeev, V.I, Shevtsov, M.B, Abramchik, Y.A, Mikheeva, O.O, Kostromina, M.A, Lykoshin, D.D, Zayats, E.A, Zavriev, S.K, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural plasticity and thermal stability of the histone-like protein from Spiroplasma melliferum are due to phenylalanine insertions into the conservative scaffold.
J.Biomol.Struct.Dyn., 36, 2018
|
|
7NY0
 
 | |
7PO7
 
 | | Phosphoglycolate phosphatase from Mus musculus | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Schloetzer, J, Schindelin, H, Fratz, S. | | Deposit date: | 2021-09-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Glycolytic flux control by drugging phosphoglycolate phosphatase.
Nat Commun, 13, 2022
|
|
7POE
 
 | | Phosphoglycolate Phosphatase with Inhibitor CP1 | | Descriptor: | 2-[[4-[4-[(2-carboxyphenyl)carbamoyl]phenoxy]phenyl]carbonylamino]benzoic acid, GLYCEROL, Glycerol-3-phosphate phosphatase, ... | | Authors: | Schloetzer, J, Fratz, S, Schindelin, H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Glycolytic flux control by drugging phosphoglycolate phosphatase.
Nat Commun, 13, 2022
|
|
3QYO
 
 | | Sensitivity of receptor internal motions to ligand binding affinity and kinetic off-rate | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Collins, E.J, Lee, A.L, Carroll, M.J, Mauldin, R.V, Gromova, A.V, Singleton, S.F. | | Deposit date: | 2011-03-03 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Evidence for dynamics in proteins as a mechanism for ligand dissociation.
Nat.Chem.Biol., 8, 2012
|
|
7QG7
 
 | | SARS-CoV-2 macrodomain Nsp3b bound to the remdesivir nucleoside GS-441524 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolane-2-carbonitrile, 1,2-ETHANEDIOL, Papain-like protease nsp3 | | Authors: | Wollenhaupt, J, Linhard, V, Sreeramulu, S, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2021-12-07 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Binding Adaptation of GS-441524 Diversifies Macro Domains and Downregulates SARS-CoV-2 de-MARylation Capacity.
J.Mol.Biol., 434, 2022
|
|
3R35
 
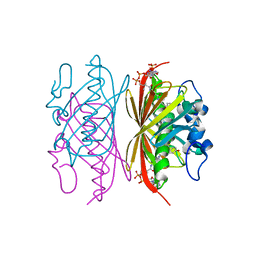 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase mutant E73D complexed with 4-hydroxyphenacyl CoA | | Descriptor: | 4-HYDROXYPHENACYL COENZYME A, 4-hydroxybenzoyl-CoA thioesterase | | Authors: | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
7Q9S
 
 | | Crystal structure of PDE6D KRas peptide complex with Compound-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3-(1~{H}-pyrazol-5-yl)imidazo[1,2-a]pyridine, DIMETHYL SULFOXIDE, ... | | Authors: | Yelland, T, Ismail, S. | | Deposit date: | 2021-11-14 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Stabilization of the RAS:PDE6D Complex Is a Novel Strategy to Inhibit RAS Signaling.
J.Med.Chem., 65, 2022
|
|
3TGI
 
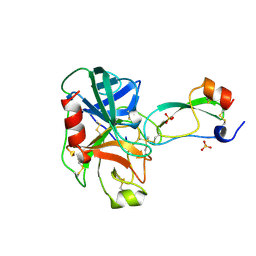 | | WILD-TYPE RAT ANIONIC TRYPSIN COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR (BPTI) | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, CALCIUM ION, SULFATE ION, ... | | Authors: | Pasternak, A, Ringe, D, Hedstrom, L. | | Deposit date: | 1998-07-15 | | Release date: | 1998-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of Anionic and Cationic Trypsinogens: The Anionic Activation Domain is More Flexible in Solution and Differs in its Mode of Bpti Binding in the Crystal Structure
Protein Sci., 8, 1999
|
|
7QJK
 
 | | Crystal structure of PDE6D in complex with Compound-2 | | Descriptor: | 1,2-ETHANEDIOL, N-(phenylmethyl)pyridin-2-amine, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, ... | | Authors: | Yelland, T, Ismail, S. | | Deposit date: | 2021-12-16 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Stabilization of the RAS:PDE6D Complex Is a Novel Strategy to Inhibit RAS Signaling.
J.Med.Chem., 65, 2022
|
|
7NT4
 
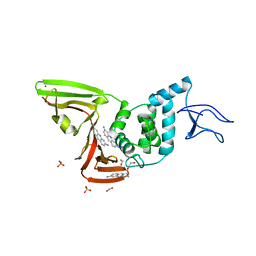 | | X-ray structure of SCoV2-PLpro in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 3, PROFLAVIN, ... | | Authors: | Napolitano, V, Mourao, A, Bostock, M, Matsuda, A, Czarna, A, Popowicz, G.M. | | Deposit date: | 2021-03-09 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Acriflavine, a clinically approved drug, inhibits SARS-CoV-2 and other betacoronaviruses.
Cell Chem Biol, 29, 2022
|
|
3TGJ
 
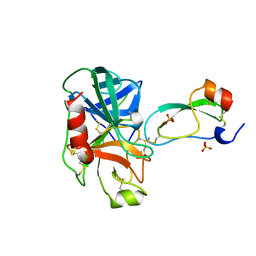 | | S195A TRYPSINOGEN COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR (BPTI) | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, CALCIUM ION, SULFATE ION, ... | | Authors: | Pasternak, A, Ringe, D, Hedstrom, L. | | Deposit date: | 1998-07-16 | | Release date: | 1998-12-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparison of Anionic and Cationic Trypsinogens: The Anionic Activation Domain is More Flexible in Solution and Differs in its Mode of Bpti Binding in the Crystal Structure
Protein Sci., 8, 1999
|
|
7NTQ
 
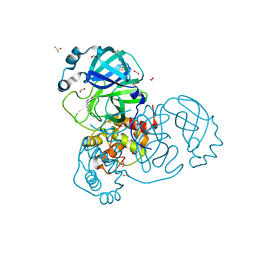 | | Crystal structure of the SARS-CoV-2 Main Protease complexed with N-(pyridin-3-ylmethyl)thioformamide | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Dupre, E, Villeret, V, Hanoulle, X. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Novel dithiocarbamates selectively inhibit 3CL protease of SARS-CoV-2 and other coronaviruses.
Eur.J.Med.Chem., 250, 2023
|
|
3RFV
 
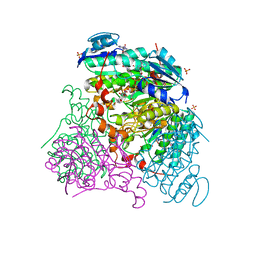 | | Crystal structure of Uronate dehydrogenase from Agrobacterium tumefaciens complexed with NADH and product | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-galactaro-1,5-lactone, PHOSPHATE ION, ... | | Authors: | Parkkinen, T, Rouvinen, J. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Uronate Dehydrogenase from Agrobacterium tumefaciens.
J.Biol.Chem., 286, 2011
|
|
7QDW
 
 | |
3RS3
 
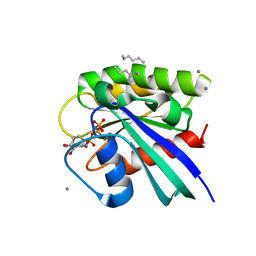 | | H-Ras soaked in neat hexane: 1 of 10 in MSCS set | | Descriptor: | CALCIUM ION, GTPase HRas, HEXANE, ... | | Authors: | Mattos, C, Buhrman, G, Kearney, B. | | Deposit date: | 2011-05-02 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Analysis of Binding Site Hot Spots on the Surface of Ras GTPase.
J.Mol.Biol., 413, 2011
|
|
3RSL
 
 | | H-Ras soaked in 90% R,S,R-bisfuranol: one of 10 in MSCS set | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-ol, CALCIUM ION, GTPase HRas, ... | | Authors: | Mattos, C, Buhrman, G, Kearney, B. | | Deposit date: | 2011-05-02 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysis of Binding Site Hot Spots on the Surface of Ras GTPase.
J.Mol.Biol., 413, 2011
|
|
3R85
 
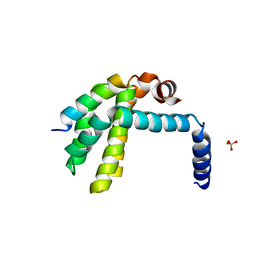 | | Crystal structure of human SOUL BH3 domain in complex with Bcl-xL | | Descriptor: | Bcl-2-like protein 1, Heme-binding protein 2, SULFATE ION | | Authors: | Ambrosi, E.K, Capaldi, S, Bovi, M, Saccomani, G, Perduca, M, Monaco, H.L. | | Deposit date: | 2011-03-23 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural changes in the BH3 domain of SOUL protein upon interaction with the anti-apoptotic protein Bcl-xL.
Biochem.J., 438, 2011
|
|
3RDJ
 
 | | Rat PKC C2 domain Apo | | Descriptor: | Protein kinase C alpha type | | Authors: | Li, P. | | Deposit date: | 2011-04-01 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pb2+ as modulator of protein-membrane interactions.
J.Am.Chem.Soc., 133, 2011
|
|
3R8K
 
 | | Crystal structure of human SOUL protein (hexagonal form) | | Descriptor: | Heme-binding protein 2 | | Authors: | Ambrosi, E.K, Capaldi, S, Bovi, M, Saccomani, G, Perduca, M, Monaco, H.L. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural changes in the BH3 domain of SOUL protein upon interaction with the anti-apoptotic protein Bcl-xL.
Biochem.J., 438, 2011
|
|
