6LFD
 
 | |
6L7M
 
 | |
2ICG
 
 | |
6L7Y
 
 | | Crystal structure of Cet1 from Trypanosoma cruzi in complex with #466 ligand. | | Descriptor: | 3,4,6,7-tetrahydroacridine-1,8(2H,5H)-dione, SULFATE ION, mRNA_triPase domain-containing protein | | Authors: | Kuwabara, N, Ho, K. | | Deposit date: | 2019-11-03 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structures of the RNA triphosphatase fromTrypanosoma cruziprovide insights into how it recognizes the 5'-end of the RNA substrate.
J.Biol.Chem., 295, 2020
|
|
6L84
 
 | | Complex of DNA polymerase IV and D-DNA duplex | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*C)-3'), DNA (5'-D(P*CP*GP*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Chung, H.S, An, J, Hwang, D. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | The crystal structure of a natural DNA polymerase complexed with mirror DNA.
Chem.Commun.(Camb.), 56, 2020
|
|
2ICQ
 
 | | urate oxidase under 2.0 MPa pressure of nitrous oxide | | Descriptor: | 8-AZAXANTHINE, CYSTEINE, NITROUS OXIDE, ... | | Authors: | Colloc'h, N, Sopkova-de Oliveira Santos, J, Retailleau, P, Prange, T, Abraini, J.H. | | Deposit date: | 2006-09-13 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein Crystallography under Xenon and Nitrous Oxide Pressure: Comparison with In Vivo Pharmacology Studies and Implications for the Mechanism of Inhaled Anesthetic Action
Biophys.J., 92, 2007
|
|
2IF7
 
 | | Crystal Structure of NTB-A | | Descriptor: | CALCIUM ION, CHLORIDE ION, SLAM family member 6 | | Authors: | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2006-09-20 | | Release date: | 2006-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | NTB-A Receptor Crystal Structure: Insights into Homophilic Interactions in the Signaling Lymphocytic Activation Molecule Receptor Family.
Immunity, 25, 2006
|
|
6L8E
 
 | | Crystal structure of heterohexameric YoeB-YefM complex bound to 26bp-DNA | | Descriptor: | DNA (26-mer), YefM Antitoxin, YoeB toxin | | Authors: | Yue, J, Xue, L. | | Deposit date: | 2019-11-06 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Distinct oligomeric structures of the YoeB-YefM complex provide insights into the conditional cooperativity of type II toxin-antitoxin system.
Nucleic Acids Res., 48, 2020
|
|
6LG1
 
 | | Crystal structure of LpCGTa in complex with UDP | | Descriptor: | LpCGTa, URIDINE-5'-DIPHOSPHATE | | Authors: | Gao, H.M, Yun, C.H. | | Deposit date: | 2019-12-04 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.047 Å) | | Cite: | Dissection of the general two-step di- C -glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LGL
 
 | | The atomic structure of varicella-zoster virus A-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
6KZZ
 
 | | Crystal structure of E.coli DNA gyrase B in complex with 2-oxo-1,2-dihydroquinoline derivative | | Descriptor: | 4-[[8-(methylamino)-2-oxidanylidene-1~{H}-quinolin-3-yl]carbonylamino]benzoic acid, DNA gyrase subunit B | | Authors: | Mima, M, Takeuchi, T, Ushiyama, F. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation.
Acs Omega, 5, 2020
|
|
6L8S
 
 | | High resolution crystal structure of crustacean hemocyanin. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Masuda, T, Mikami, B, Baba, S. | | Deposit date: | 2019-11-07 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The high-resolution crystal structure of lobster hemocyanin shows its enzymatic capability as a phenoloxidase.
Arch.Biochem.Biophys., 688, 2020
|
|
6LA6
 
 | |
6LAS
 
 | | the wildtype SAM-VI riboswitch bound to SAM | | Descriptor: | RNA (55-MER), S-ADENOSYLMETHIONINE, U1 small nuclear ribonucleoprotein A | | Authors: | Ren, A, Sun, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
6L0A
 
 | | Crystal structure of dihydroorotase in complex with malate at pH7 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2021-12-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Analysis of Saccharomyces cerevisiae Dihydroorotase Reveals Molecular Insights into the Tetramerization Mechanism
Molecules, 2021
|
|
6L0W
 
 | | Structure of RLD2 BRX domain bound to LZY3 CCL motif | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, NGR2, ... | | Authors: | Hirano, Y, Futrutani, M, Nishimura, T, Taniguchi, M, Morita, M.T, Hakoshima, T. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control.
Nat Commun, 11, 2020
|
|
2II7
 
 | | Anabaena sensory rhodopsin transducer | | Descriptor: | Anabaena sensory rhodopsin transducer protein | | Authors: | Vogeley, L. | | Deposit date: | 2006-09-27 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the anabaena sensory rhodopsin transducer.
J.Mol.Biol., 367, 2007
|
|
6L14
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-chloranyl-10-[3-[(3~{S})-piperidin-3-yl]propyl]phenoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6LB6
 
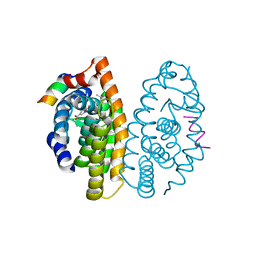 | | Crystal structure of dimeric RXR-LBD complexed with partial agonist NEt-4IB and TIF2 co-activator | | Descriptor: | 6-[ethyl-[4-(2-methylpropoxy)-3-propan-2-yl-phenyl]amino]pyridine-3-carboxylic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Imai, D, Numoto, N, Nakano, S, Kakuta, H, Ito, N. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the full and partial agonist activities of retinoid X receptor alpha ligands with an iso-butoxy and an isopropyl group
Biochem.Biophys.Res.Commun., 734, 2024
|
|
6LBE
 
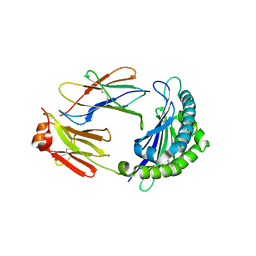 | |
2IFC
 
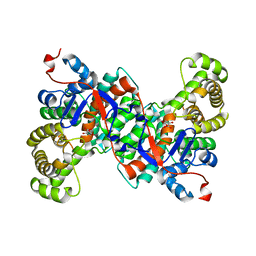 | |
6LBP
 
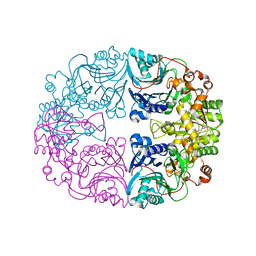 | | Structure of the Glutamine Phosphoribosylpyrophosphate Amidotransferase from Arabidopsis thaliana | | Descriptor: | Amidophosphoribosyltransferase 2, chloroplastic, IRON/SULFUR CLUSTER | | Authors: | Yi, Z, Cao, X, Han, F, Feng, Y. | | Deposit date: | 2019-11-14 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.065 Å) | | Cite: | Crystal Structure of the Chloroplastic Glutamine Phosphoribosylpyrophosphate Amidotransferase GPRAT2 FromArabidopsis thaliana.
Front Plant Sci, 11, 2020
|
|
6LCC
 
 | |
6LCQ
 
 | | Crystal structure of rice defensin OsAFP1 | | Descriptor: | Defensin-like protein CAL1, PHOSPHATE ION | | Authors: | Ochiai, A, Ogawa, K, Fukuda, M, Suzuki, M, Ito, K, Tanaka, T, Sagehashi, Y, Taniguchi, M. | | Deposit date: | 2019-11-19 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of rice defensin OsAFP1 and molecular insight into lipid-binding.
J.Biosci.Bioeng., 130, 2020
|
|
6LB3
 
 | | Crystal structure of PA4674 in complex with its operator DNA (18bp) from Pseudomonas aeruginosa | | Descriptor: | DNA (5'-D(P*AP*CP*CP*CP*TP*TP*AP*AP*CP*GP*TP*TP*AP*AP*GP*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*CP*TP*TP*AP*AP*CP*GP*TP*TP*AP*AP*GP*GP*GP*T)-3'), HTH cro/C1-type domain-containing protein, ... | | Authors: | Liu, Y, Zhang, H, Gao, Z, Dong, Y. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Crystal structure of PA4674 in complex with its operator DNA (18bp) from Pseudomonas aeruginosa
To Be Published
|
|
