6L6M
 
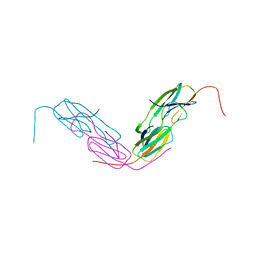 | | HSP18.5 from E. histolytica | | Descriptor: | Heat shock protein hsp20 family putative | | Authors: | Kurre, D, Suguna, K. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.28000379 Å) | | Cite: | Network of Entamoeba histolytica HSP18.5 dimers formed by two overlapping [IV]-X-[IV] motifs.
Proteins, 2021
|
|
6L6Y
 
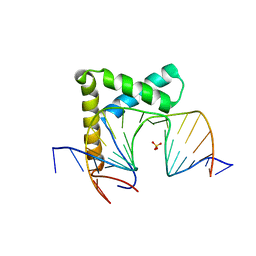 | |
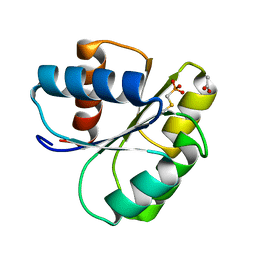
6L7X
 
 | | Crystal structure of Cet1 from Trypanosoma cruzi in complex with #951 ligand | | Descriptor: | 1,3-dimethyl-7-propyl-purine-2,6-dione, SULFATE ION, mRNA_triPase domain-containing protein | | Authors: | Kuwabara, N, Ho, K. | | Deposit date: | 2019-11-03 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structures of the RNA triphosphatase fromTrypanosoma cruziprovide insights into how it recognizes the 5'-end of the RNA substrate.
J.Biol.Chem., 295, 2020
|
|
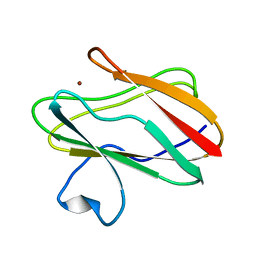
2IDM
 
 | | 2.00 A Structure of T87I/Y106W Phosphono-CheY | | Descriptor: | ACETATE ION, Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-15 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
2IDS
 
 | | Structure of M98A mutant of amicyanin, Cu(I) | | Descriptor: | Amicyanin, COPPER (I) ION | | Authors: | Carrell, C.J, Ma, J.K, Antholine, W, Hosler, J.P, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-09-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Generation of Novel Copper Sites by Mutation of the Axial Ligand of Amicyanin. Atomic Resolution Structures and Spectroscopic Properties
Biochemistry, 46, 2007
|
|
6KZG
 
 | |
2IDX
 
 | |
6KZV
 
 | |
6L04
 
 | | Crystal structure of uPA_H99Y in complex with 31F | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-N-carbamimidoyl-6-(2,4-dimethoxypyrimidin-5-yl)pyrazine-2-carboxamide, Urokinase-type plasminogen activator | | Authors: | Buckley, B, Jiang, L.G, Huang, M.D, Kelso, M, Ranson, M. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of uPA_H99Y in complex with 31F
To Be Published
|
|
6L0G
 
 | | Crystal structure of dihydroorotase in complex with malate at pH6 from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil.
Biochem.Biophys.Res.Commun., 551, 2021
|
|
2ITD
 
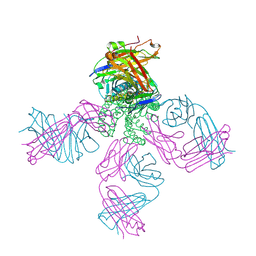 | | Potassium Channel KcsA-Fab complex in Barium Chloride | | Descriptor: | BARIUM ION, Voltage-gated potassium channel, antibody Fab fragment heavy chain, ... | | Authors: | Lockless, S.W, Zhou, M, MacKinnon, R. | | Deposit date: | 2006-10-19 | | Release date: | 2007-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Thermodynamic Properties of Selective Ion Binding in a K(+) Channel.
Plos Biol., 5, 2007
|
|
6L82
 
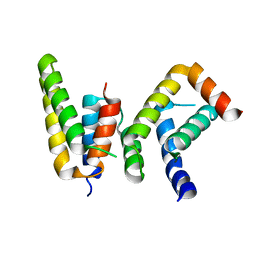 | | Crystal structure of Chaetomium GCP5 N-terminus and Mozart1 | | Descriptor: | Mozart1, Spindle pole body component | | Authors: | Huang, T.L, Wang, H.J, Wang, S.W, Hsia, K.C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.24103618 Å) | | Cite: | Promiscuous Binding of Microprotein Mozart1 to gamma-Tubulin Complex Mediates Specific Subcellular Targeting to Control Microtubule Array Formation.
Cell Rep, 31, 2020
|
|
6L88
 
 | |
6L8K
 
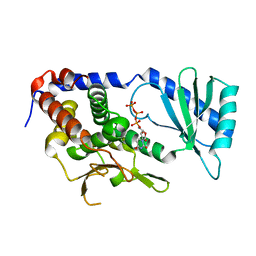 | | Structure of URT1 in complex with UTP | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, UTP:RNA uridylyltransferase 1 | | Authors: | Lingru, Z. | | Deposit date: | 2019-11-06 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Crystal structure of Arabidopsis terminal uridylyl transferase URT1.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
6L8T
 
 | |
6L13
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-chloranyl-10-(2-piperidin-4-ylethyl)phenoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L1F
 
 | |
6L9X
 
 | | Xenons in frog EPDR1 | | Descriptor: | Ependymin-related 1, XENON | | Authors: | Park, S. | | Deposit date: | 2019-11-11 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | De novo Phasing Xenons Observed in the Frog Ependymin-Related Protein
Crystals, 10, 2020
|
|
6LA4
 
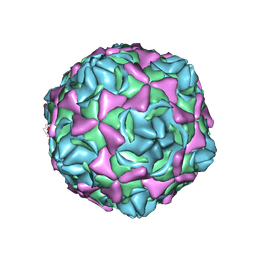 | | Cryo-EM structure of full echovirus 11 particle at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-11 | | Release date: | 2020-10-07 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LAH
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with lysophosphatidylcholine | | Descriptor: | (2R)-2,3-dihydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shima, Y, Morita, D. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of lysophospholipid-bound MHC class I molecules.
J.Biol.Chem., 295, 2020
|
|
6LAZ
 
 | | the wildtype SAM-VI riboswitch bound to a N-mustard SAM analog M1 | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(2-hydroxyethyl)amino]-2-azaniumyl-butanoate, MAGNESIUM ION, RNA (55-MER), ... | | Authors: | Ren, A, Sun, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
6L1Q
 
 | | Crystal structure of AfCbbQ2, a MoxR AAA+-ATPase and CbbQO-type Rubisco activase from Acidithiobacillus ferrooxidans | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CbbQ protein, PHOSPHATE ION | | Authors: | Ye, F.Z, Tsai, Y.C.C, Mueller-Cajar, O, Gao, Y.G. | | Deposit date: | 2019-09-30 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the mechanism and regulation of the CbbQO-type Rubisco activase, a MoxR AAA+ ATPase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L2L
 
 | |
6L2R
 
 | | IlvC, a ketol-acid reductoisomerase, from Streptococcus pneumoniae_E195S | | Descriptor: | Ketol-acid reductoisomerase (NADP(+)) | | Authors: | Gyuhee, K, Donghyuk, S, Sumin, L, Jaesook, Y, Sangho, L. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of IlvC, a Ketol-Acid Reductoisomerase, from Streptococcus Pneumoniae.
Crystals, 9, 2019
|
|
6L36
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
