6V1X
 
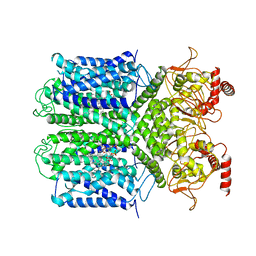 | | Cryo-EM Structure of the Hyperpolarization-Activated Potassium Channel KAT1: Tetramer | | Descriptor: | (2S)-1-(nonanoyloxy)-3-(phosphonooxy)propan-2-yl tetradecanoate, (3beta,5beta,14beta,17alpha)-cholestan-3-ol, Potassium channel KAT1 | | Authors: | Clark, M.D, Contreras, G.F, Shen, R, Perozo, E. | | Deposit date: | 2019-11-21 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Electromechanical coupling in the hyperpolarization-activated K + channel KAT1.
Nature, 583, 2020
|
|
6UPV
 
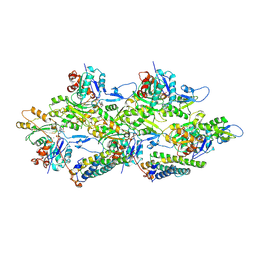 | | Alpha-E-catenin ABD-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mei, L, Alushin, G.M. | | Deposit date: | 2019-10-18 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular mechanism for direct actin force-sensing by alpha-catenin.
Elife, 9, 2020
|
|
3IK6
 
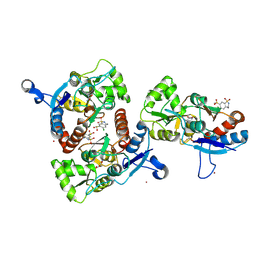 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, chlorothiazide | | Descriptor: | 6-chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ptak, C.P, Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
6LP9
 
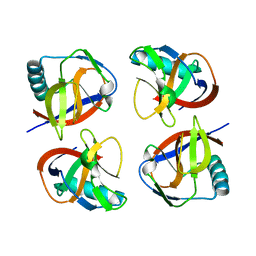 | | the protein of cat virus | | Descriptor: | nsp1 protein | | Authors: | Shen, Z, Peng, G.Q. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural and Biological Basis of Alphacoronavirus nsp1 Associated with Host Proliferation and Immune Evasion.
Viruses, 12, 2020
|
|
4Q4I
 
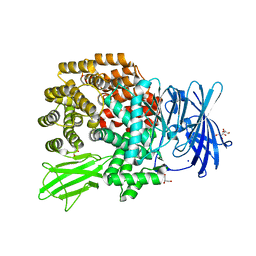 | | Crystal structure of E.coli aminopeptidase N in complex with amastatin | | Descriptor: | Amastatin, Aminopeptidase N, GLYCEROL, ... | | Authors: | Reddi, R, Ganji, R.J, Addlagatta, A. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis for the inhibition of M1 family aminopeptidases by the natural product actinonin: Crystal structure in complex with E. coli aminopeptidase N.
Protein Sci., 24, 2015
|
|
6UVS
 
 | |
3ITM
 
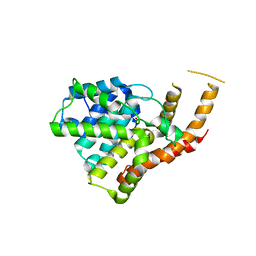 | | Catalytic domain of hPDE2A | | Descriptor: | ZINC ION, cGMP-dependent 3',5'-cyclic phosphodiesterase | | Authors: | Pandit, J. | | Deposit date: | 2009-08-28 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4QAA
 
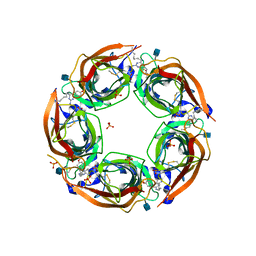 | | X-RAY STRUCTURE OF ACETYLCHOLINE BINDING PROTEIN (ACHBP) IN COMPLEX WITH 6-(4-Methoxyphenyl)-N4-octylpyrimidine-2,4-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(4-methoxyphenyl)-N~4~-octylpyrimidine-2,4-diamine, Acetylcholine-binding protein, ... | | Authors: | Kaczanowska, K, Harel, M, Radic, Z, Changeux, J.-P, Finn, M.G, Taylor, P. | | Deposit date: | 2014-05-03 | | Release date: | 2014-07-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for cooperative interactions of substituted 2-aminopyrimidines with the acetylcholine binding protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8DB5
 
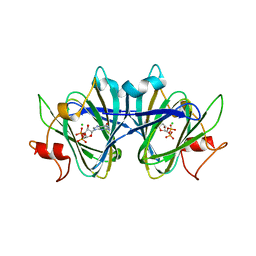 | | Crystal structure of the GDP-D-glycero-4-keto-d-lyxo-heptose-3,5-epimerase from Campylobacter jejuni, serotype HS:15 | | Descriptor: | CHLORIDE ION, GDP-D-glycero-4-keto-d-lyxo-heptose-3,5-epimerase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Thoden, J.B, Ghosh, M.K, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-14 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
6ANV
 
 | | Crystal structure of anti-CRISPR protein AcrF1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, H, Patel, D.J. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.265 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
8DCO
 
 | | Crystal structure of the GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase from Campylobacter jejuni, serotype HS:42 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase, ... | | Authors: | Thoden, J.B, Xiang, D.F, Ghosh, M.K, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
6ATJ
 
 | | RECOMBINANT HORSERADISH PEROXIDASE C COMPLEX WITH FERULIC ACID | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, CALCIUM ION, PEROXIDASE C1A, ... | | Authors: | Henriksen, A, Smith, A.T, Gajhede, M. | | Deposit date: | 1999-04-23 | | Release date: | 2000-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of the horseradish peroxidase C-ferulic acid complex and the ternary complex with cyanide suggest how peroxidases oxidize small phenolic substrates.
J.Biol.Chem., 274, 1999
|
|
8DCL
 
 | | Crystal structure of the GDP-D-glycero-4-keto-D-lyxo-heptose-3-epimerase from campylobacter jejuni, serotype HS:23/36 | | Descriptor: | 1,2-ETHANEDIOL, GDP-D-glycero-4-keto-D-lyxo-heptose-3-epimerase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Ghosh, M.K, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-16 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
8D14
 
 | | Helical ADP-Pi-F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
8D18
 
 | | Straight ADP-F-actin 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
7N9F
 
 | | Structure of the in situ yeast NPC | | Descriptor: | Dynein light chain 1, cytoplasmic, Nucleoporin 145c, ... | | Authors: | Villa, E, Singh, D, Ludtke, S.J, Akey, C.W, Rout, M.P, Echeverria, I, Suslov, S. | | Deposit date: | 2021-06-17 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (37 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
8DAK
 
 | | Crystal structure of the GDP-D-glycero-4-keto-d-lyxo-heptose-3-epimerase from Campylobacter jejuni, serotype HS:3 | | Descriptor: | CHLORIDE ION, GDP-D-glycero-4-keto-d-lyxo-heptose-3-epimerase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Ghosh, M.K, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-13 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
7Q60
 
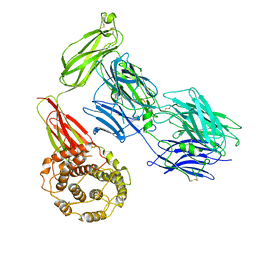 | |
7Q5Z
 
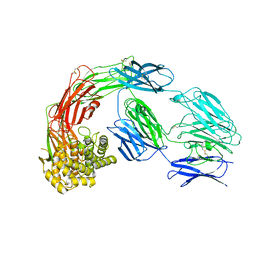 | |
5IPX
 
 | | Structure of ORF49 from KSHV | | Descriptor: | ORF49 protein, SULFATE ION | | Authors: | Hew, K, Nordlund, P. | | Deposit date: | 2016-03-10 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Open Reading Frame 49 Protein Encoded by Kaposi's Sarcoma-Associated Herpesvirus.
J. Virol., 91, 2017
|
|
8D15
 
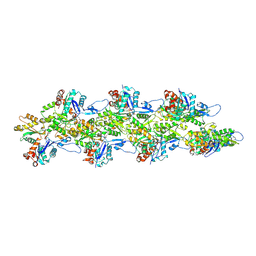 | | Bent ADP-F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
7Q1Y
 
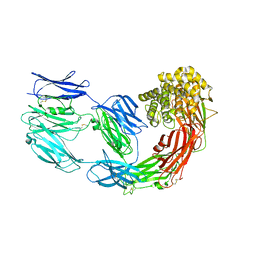 | | X-ray structure of human A2ML1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin-like protein 1, ... | | Authors: | Andersen, G.R, Zarantonello, A, Enghild, J.J, Nielsen, N.S. | | Deposit date: | 2021-10-22 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Cryo-EM structures of human A2ML1 elucidate the protease-inhibitory mechanism of the A2M family.
Nat Commun, 13, 2022
|
|
4Q85
 
 | | YcaO with Non-hydrolyzable ATP (AMPCPP) Bound | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, Ribosomal protein S12 methylthiotransferase accessory factor YcaO | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2014-04-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis.
Nat.Chem.Biol., 10, 2014
|
|
7Q62
 
 | |
7PWG
 
 | | Cryo-EM structure of large subunit of Giardia lamblia ribosome at 2.7 A resolution | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L18a, 60S ribosomal protein L27, ... | | Authors: | Hiregange, D.G, Rivalta, A, Bose, T, Breiner-Goldstein, E, Samiya, S, Cimicata, G, Kulakova, L, Zimmerman, E, Bashan, A, Herzberg, O, Yonath, A. | | Deposit date: | 2021-10-06 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Cryo-EM structure of the ancient eukaryotic ribosome from the human parasite Giardia lamblia.
Nucleic Acids Res., 50, 2022
|
|
