3TGR
 
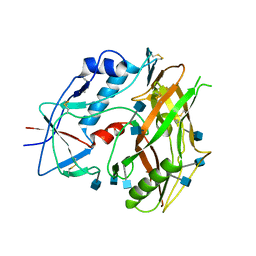 | | Crystal structure of unliganded HIV-1 clade C strain C1086 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade C1086 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TH9
 
 | | Crystal Structure of HIV-1 Protease Mutant Q7K V32I L63I with a cyclic sulfonamide inhibitor | | Descriptor: | Gag-Pol polyprotein, tert-butyl {(2S,3R)-4-[(4S)-7-fluoro-4-methyl-1,1-dioxido-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}carbamate | | Authors: | Orth, P. | | Deposit date: | 2011-08-18 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Design, Synthesis, and X-ray Crystallographic Analysis of a Novel Class of HIV-1 Protease Inhibitors.
J.Med.Chem., 54, 2011
|
|
3SQB
 
 | | Structure of the major type 1 pilus subunit FimA bound to the FimC chaperone | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chaperone protein fimC, ... | | Authors: | Scharer, M.A, Eidam, O, Grutter, M.G, Glockshuber, R, Capitani, G. | | Deposit date: | 2011-07-05 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Quality control of disulfide bond formation in pilus subunits by the chaperone FimC.
Nat.Chem.Biol., 8, 2012
|
|
3SRR
 
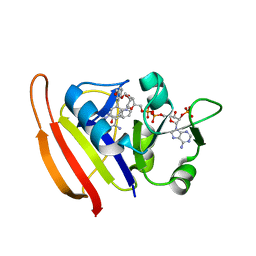 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | 3-(2,4-diamino-6-methylquinazolin-7-yl)-4-ethoxybenzaldehyde, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3TGT
 
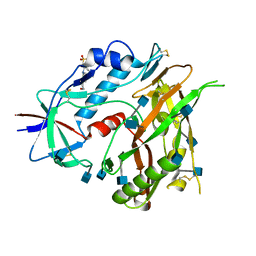 | | Crystal structure of unliganded HIV-1 clade A/E strain 93TH057 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 clade A/E 93TH057 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3THF
 
 | |
3SSD
 
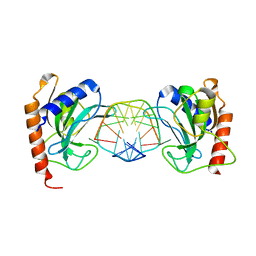 | | DNA binding domain of restriction endonuclease bound to DNA | | Descriptor: | 5-methylcytosine-specific restriction enzyme B, DNA (5'-D(*A*GP*CP*TP*AP*CP*CP*GP*GP*TP*CP*TP*C)-3'), DNA (5'-D(*T*GP*AP*GP*AP*(5CM)P*CP*GP*GP*TP*AP*GP*C)-3') | | Authors: | Sukackaite, R, Grazulis, S, Siksnys, V. | | Deposit date: | 2011-07-08 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The recognition domain of the methyl-specific endonuclease McrBC flips out 5-methylcytosine.
Nucleic Acids Res., 40, 2012
|
|
3SSP
 
 | |
3STK
 
 | |
3SQY
 
 | |
3STX
 
 | |
3SR5
 
 | |
3SRW
 
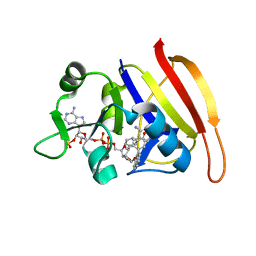 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | 7-(2-ethoxynaphthalen-1-yl)-6-methylquinazoline-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SSV
 
 | |
3STY
 
 | |
3SUD
 
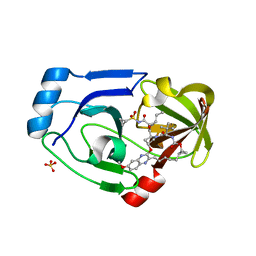 | | Crystal structure of NS3/4A protease in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-di oxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadec ino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SXD
 
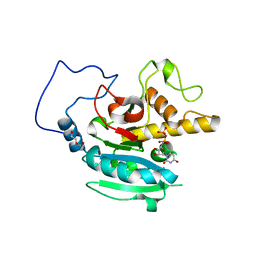 | | Crystal structure of BBBB+UDP+Gal with MPD as the cryoprotectant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Johal, A.R, Evans, S.V. | | Deposit date: | 2011-07-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Sequence-dependent effects of cryoprotectants on the active sites of the human ABO(H) blood group A and B glycosyltransferases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SVD
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: bromide complex | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Determination of engineered chloride-binding site structures in fluorescent proteins reveals principles of halide recognition
To be Published
|
|
3SYQ
 
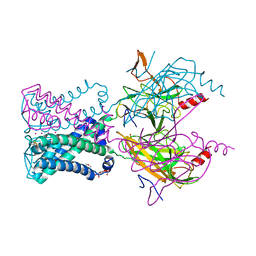 | | Crystal structure of the G protein-gated inward rectifier K+ channel GIRK2 (Kir3.2) R201A mutant in complex with PIP2 | | Descriptor: | G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Whorton, M.R, MacKinnon, R. | | Deposit date: | 2011-07-18 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Crystal Structure of the Mammalian GIRK2 K(+) Channel and Gating Regulation by G Proteins, PIP(2), and Sodium.
Cell(Cambridge,Mass.), 147, 2011
|
|
3SY6
 
 | |
3SZ9
 
 | |
3T0J
 
 | |
3T1H
 
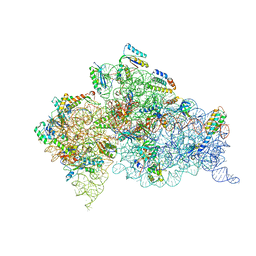 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA lysine 3 (tRNALys3) bound to an mRNA with an AAA-codon in the A-site and Paromomycin | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Murphy, F.V, Vendeix, F.A.P, Cantara, W, Leszczynska, G, Gustilo, E.M, Sproat, B, Malkiewicz, A.A.P, Agris, P.F. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Human tRNA(Lys3)(UUU) Is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-Enol Tautomerism.
J.Mol.Biol., 416, 2012
|
|
3T4V
 
 | | Crystal Structure of AlkB in complex with Fe(III) and N-Oxalyl-S-(2-napthalenemethyl)-L-cysteine | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase AlkB, FE (III) ION, GLYCEROL, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2011-07-26 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Dynamic combinatorial mass spectrometry leads to inhibitors of a 2-oxoglutarate-dependent nucleic Acid demethylase.
J.Med.Chem., 55, 2012
|
|
3T4P
 
 | |
