2VRM
 
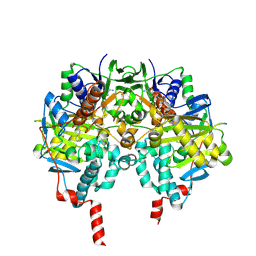 | | Structure of human MAO B in complex with phenyethylhydrazine | | Descriptor: | AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE, PHENYLETHANE | | Authors: | Binda, C, Wang, J, Li, M, Hubalek, F, Mattevi, A, Edmondson, D.E. | | Deposit date: | 2008-04-09 | | Release date: | 2008-04-22 | | Last modified: | 2014-01-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Studies of Arylalkylhydrazine Inhibition of Human Monoamine Oxidases a and B
Biochemistry, 47, 2008
|
|
2VRL
 
 | | Structure of human MAO B in complex with benzylhydrazine | | Descriptor: | AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE, TOLUENE | | Authors: | Binda, C, Wang, J, Li, M, Hubalek, F, Mattevi, A, Edmondson, D.E. | | Deposit date: | 2008-04-09 | | Release date: | 2008-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mechanistic Studies of Arylalkylhydrazine Inhibition of Human Monoamine Oxidases a and B
Biochemistry, 47, 2008
|
|
3NBE
 
 | | Clitocybe nebularis ricin B-like lectin (CNL) in complex with N,N'-diacetyllactosediamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-2-azidoethyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Ricin B-like lectin, SULFATE ION | | Authors: | Renko, M, Pohleven, J, Sabotic, J, Kos, J, Turk, D. | | Deposit date: | 2010-06-03 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Bivalent carbohydrate binding is required for biological activity of Clitocybe nebularis lectin (CNL), the N,N'-diacetyllactosediamine (GalNAc beta 1-4GlcNAc, LacdiNAc)-specific lectin from basidiomycete C. nebularis
J.Biol.Chem., 287, 2012
|
|
4MR8
 
 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist CGP35348 | | Descriptor: | (S)-(3-aminopropyl)(diethoxymethyl)phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-17 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4MR7
 
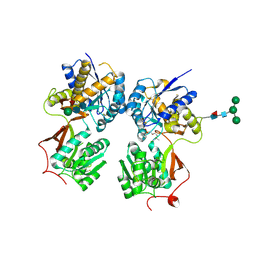 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist CGP54626 | | Descriptor: | (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-17 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4MS1
 
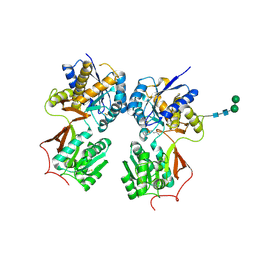 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist CGP46381 | | Descriptor: | (S)-(3-aminopropyl)(cyclohexylmethyl)phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
3OE9
 
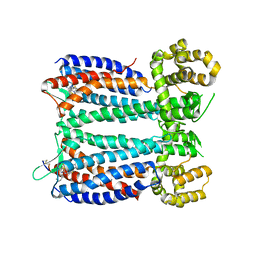 | | Crystal structure of the chemokine CXCR4 receptor in complex with a small molecule antagonist IT1t in P1 spacegroup | | Descriptor: | (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, Lysozyme Chimera | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
6BXP
 
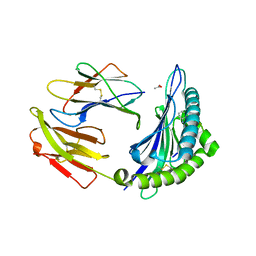 | |
5T8F
 
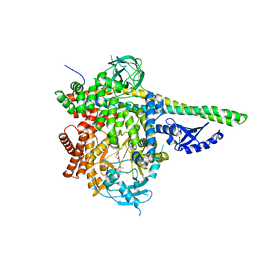 | | p110delta/p85alpha with taselisib (GDC-0032) | | Descriptor: | 2-methyl-2-(4-{2-[3-methyl-1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-1H-pyrazol-1-yl)propanamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Moertl, M, Steinbacher, S, Eigenbrot, C. | | Deposit date: | 2016-09-07 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure-Based Design of Tricyclic NF-kappa B Inducing Kinase (NIK) Inhibitors That Have High Selectivity over Phosphoinositide-3-kinase (PI3K).
J. Med. Chem., 60, 2017
|
|
5T8O
 
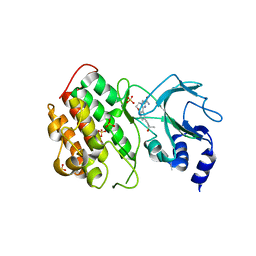 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) bound to Imidazobenzoxepin Compound 3 | | Descriptor: | 10-(3-methyl-3-oxidanyl-but-1-ynyl)-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepine-2-carboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Smith, M.A, McEwan, P, Hymowitz, S.G. | | Deposit date: | 2016-09-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-Based Design of Tricyclic NF-kappa B Inducing Kinase (NIK) Inhibitors That Have High Selectivity over Phosphoinositide-3-kinase (PI3K).
J. Med. Chem., 60, 2017
|
|
6TTU
 
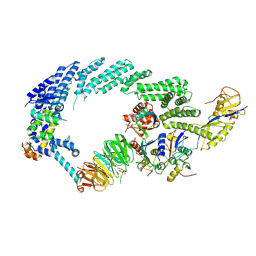 | | Ubiquitin Ligation to substrate by a cullin-RING E3 ligase at 3.7A resolution: NEDD8-CUL1-RBX1 N98R-SKP1-monomeric b-TRCP1dD-IkBa-UB~UBE2D2 | | Descriptor: | CYS-LYS-LYS-ALA-ARG-HIS-ASP-SEP-GLY, Cullin-1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly.
Nature, 578, 2020
|
|
2XHL
 
 | | Structure of a functional derivative of Clostridium botulinum neurotoxin type B | | Descriptor: | BOTULINUM NEUROTOXIN B HEAVY CHAIN, BOTULINUM NEUROTOXIN B LIGHT CHAIN, ZINC ION | | Authors: | Masuyer, G, Beard, M, Cadd, V.A, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2010-06-18 | | Release date: | 2010-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Activity of a Functional Derivative of Clostridium Botulinum Neurotoxin B.
J.Struct.Biol., 174, 2011
|
|
4M44
 
 | | Crystal structure of hemagglutinin of influenza virus B/Yamanashi/166/1998 in complex with avian-like receptor LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Ni, F, Kondrashkina, E, Wang, Q. | | Deposit date: | 2013-08-06 | | Release date: | 2013-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the divergent evolution of influenza B virus hemagglutinin.
Virology, 446, 2013
|
|
6HPF
 
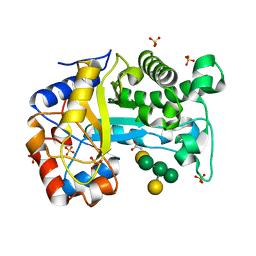 | | Structure of Inactive E165Q mutant of fungal non-CBM carrying GH26 endo-b-mannanase from Yunnania penicillata in complex with alpha-62-61-di-galactosyl-mannotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CHLORIDE ION, ... | | Authors: | von Freiesleben, P, Moroz, O.V, Blagova, E, Wiemann, M, Spodsberg, N, Agger, J.W, Davies, G.J, Wilson, K.S, Stalbrand, H, Meyer, A.S, Krogh, K.B.R.M. | | Deposit date: | 2018-09-20 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure and substrate interactions of an unusual fungal non-CBM carrying GH26 endo-beta-mannanase from Yunnania penicillata.
Sci Rep, 9, 2019
|
|
2VZ2
 
 | | Human MAO B in complex with mofegiline | | Descriptor: | (1Z)-4-(4-FLUOROPHENYL)-2-METHYLIDENEBUTAN-1-IMINE, AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bonivento, D, Mattevi, A. | | Deposit date: | 2008-07-29 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic studies of mofegiline inhibition of recombinant human monoamine oxidase B.
J. Med. Chem., 51, 2008
|
|
3PO7
 
 | | Human monoamine oxidase B in complex with zonisamide | | Descriptor: | 1-(1,2-benzoxazol-3-yl)methanesulfonamide, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, Aldeco, M, Mattevi, A, Edmondson, D.E. | | Deposit date: | 2010-11-22 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interactions of monoamine oxidases with the antiepileptic drug zonisamide: specificity of inhibition and structure of the human monoamine oxidase B complex
J.Med.Chem., 54, 2011
|
|
4NRJ
 
 | | Structure of hemagglutinin with F95Y mutation of influenza virus B/Lee/40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ HA1 CHAIN, ... | | Authors: | Ni, F, Mbawuike, I.N, Kondrashkina, E, Wang, Q. | | Deposit date: | 2013-11-26 | | Release date: | 2014-03-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The roles of hemagglutinin Phe-95 in receptor binding and pathogenicity of influenza B virus.
Virology, 450-451, 2014
|
|
3NBC
 
 | | Clitocybe nebularis ricin B-like lectin (CNL) in complex with lactose, crystallized at pH 4.4 | | Descriptor: | Ricin B-like lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Renko, M, Pohleven, J, Sabotic, J, Kos, J, Turk, D. | | Deposit date: | 2010-06-03 | | Release date: | 2011-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Bivalent carbohydrate binding is required for biological activity of Clitocybe nebularis lectin (CNL), the N,N'-diacetyllactosediamine (GalNAc beta 1-4GlcNAc, LacdiNAc)-specific lectin from basidiomycete C. nebularis
J.Biol.Chem., 287, 2012
|
|
6N0Q
 
 | | BRAF in complex with N-(4-methyl-3-(1-methyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)phenyl)-3-(trifluoromethyl)benzamide. | | Descriptor: | N-[4-methyl-3-(1-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)phenyl]-3-(trifluoromethyl)benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-11-07 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Design and Discovery ofN-(3-(2-(2-Hydroxyethoxy)-6-morpholinopyridin-4-yl)-4-methylphenyl)-2-(trifluoromethyl)isonicotinamide, a Selective, Efficacious, and Well-Tolerated RAF Inhibitor Targeting RAS Mutant Cancers: The Path to the Clinic.
J.Med.Chem., 63, 2020
|
|
5DNK
 
 | | The structure of PKMT1 from Rickettsia prowazekii in complex with AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, protein lysine methyltransferase 1 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
5HPU
 
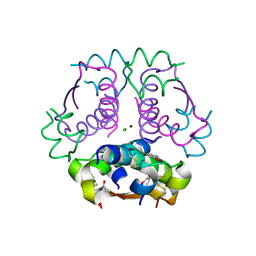 | | Insulin with proline analog HyP at position B28 in the R6 state | | Descriptor: | CHLORIDE ION, Insulin, chain A, ... | | Authors: | Lieblich, S.A, Fang, K.Y, Cahn, J.K.B, Tirrell, D.A. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 4S-Hydroxylation of Insulin at ProB28 Accelerates Hexamer Dissociation and Delays Fibrillation.
J. Am. Chem. Soc., 139, 2017
|
|
5HRQ
 
 | | Insulin with proline analog HzP at position B28 in the R6 state | | Descriptor: | CHLORIDE ION, Insulin A-Chain, Insulin B-Chain, ... | | Authors: | Lieblich, S.A, Fang, K.Y, Cahn, J.K.B, Tirrell, D.A. | | Deposit date: | 2016-01-24 | | Release date: | 2017-01-25 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 4S-Hydroxylation of Insulin at ProB28 Accelerates Hexamer Dissociation and Delays Fibrillation.
J. Am. Chem. Soc., 139, 2017
|
|
2XWJ
 
 | | Crystal Structure of Complement C3b in Complex with Factor B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3 ALPHA CHAIN, COMPLEMENT C3 BETA CHAIN, ... | | Authors: | Forneris, F, Ricklin, D, Wu, J, Tzekou, A, Wallace, R.S, Lambris, J.D, Gros, P. | | Deposit date: | 2010-11-04 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structures of C3B in Complex with Factors B and D Give Insight Into Complement Convertase Formation.
Science, 330, 2010
|
|
2XWB
 
 | | Crystal Structure of Complement C3b in complex with Factors B and D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3B ALPHA' CHAIN, ... | | Authors: | Forneris, F, Ricklin, D, Wu, J, Tzekou, A, Wallace, R.S, Lambris, J.D, Gros, P. | | Deposit date: | 2010-11-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structures of C3B in Complex with Factors B and D Give Insight Into Complement Convertase Formation.
Science, 330, 2010
|
|
1W01
 
 | |
