8V01
 
 | |
8CVD
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, succinate, and scopolamine | | Descriptor: | (1R,2R,4S,5S,7s)-9-methyl-3-oxa-9-azatricyclo[3.3.1.0~2,4~]nonan-7-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FE (II) ION, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Structure of the H6H cyclization product complex
To Be Published
|
|
6ON4
 
 | | Crystal structure of the GntR-type sialoregulator NanR from Escherichia coli, in complex with sialic acid | | Descriptor: | HTH-type transcriptional repressor NanR, N-acetyl-beta-neuraminic acid, ZINC ION, ... | | Authors: | Horne, C.R, Panjikar, S, North, R.A, Dobson, R.C.J. | | Deposit date: | 2019-04-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Control of the Escherichia coli sialoregulon by transcriptional repressor NanR.
J. Bacteriol., 195, 2013
|
|
8CVH
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the L289F H6H cyclization ferryl-mimicking complex
To Be Published
|
|
8CVA
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, succinate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FE (II) ION, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Structure of the H6H hydroxylation product complex
To Be Published
|
|
6VKT
 
 | |
8CV8
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Structure of the H6H hydroxylation reactant complex
To Be Published
|
|
6VM4
 
 | | Chloroplast ATP synthase (C2, CF1FO) | | Descriptor: | ATP synthase delta chain, chloroplastic, ATP synthase epsilon chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.08 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
8CVC
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structure mimicking the H6H cyclization ferryl complex
To Be Published
|
|
8CVB
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Structure of the H6H cyclization reactant complex
To Be Published
|
|
2VAE
 
 | | Fast maturing red fluorescent protein, DsRed.T4 | | Descriptor: | 1,2-ETHANEDIOL, RED FLUORESCENT PROTEIN | | Authors: | Strongin, D.E, Bevis, B, Khuong, N, Downing, M.E, Strack, R.L, Sundaram, K, Glick, B.S, Keenan, R.J. | | Deposit date: | 2007-08-31 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Rearrangements Near the Chromophore Influence the Maturation Speed and Brightness of Dsred Variants.
Protein Eng.Des.Sel., 20, 2007
|
|
8CVF
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Hyoscyamine 6-beta-hydroxylase, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Structure of the L289F H6H ferryl-mimicking complex
To Be Published
|
|
8UWW
 
 | |
8CVE
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure of the L289F H6H hydroxylation reactant complex
To Be Published
|
|
8UYD
 
 | |
8CV9
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Hyoscyamine 6-beta-hydroxylase, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of the H6H hydroxylation reactant complex
To Be Published
|
|
8CVG
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of the L289F H6H cyclization reactant complex
To Be Published
|
|
6OOU
 
 | | Crystal structure of HIV-1 Protease NL4-3 L89V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, NL4-3 PROTEASE, SULFATE ION | | Authors: | Henes, M, Kosovrasti, K, Lockbaum, G.J, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A, Whitfield, T.W. | | Deposit date: | 2019-04-23 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.127 Å) | | Cite: | Molecular Determinants of Epistasis in HIV-1 Protease: Elucidating the Interdependence of L89V and L90M Mutations in Resistance.
Biochemistry, 58, 2019
|
|
7K6N
 
 | | Crystal structure of PI3Kalpha selective Inhibitor 11-1575 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, tert-butyl (3S)-3-[4-(2-aminopyrimidin-5-yl)-2-(morpholin-4-yl)-5,6-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]-3-methylpyrrolidine-1-carboxylate | | Authors: | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | Deposit date: | 2020-09-21 | | Release date: | 2021-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
7W0L
 
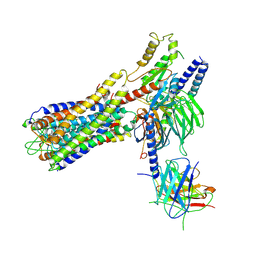 | | Cryo-EM structure of a dimeric GPCR-Gi complex with small molecule | | Descriptor: | (1R,2S)-N-[4-(2,6-dimethoxyphenyl)-5-(6-methylpyridin-2-yl)-1,2,4-triazol-3-yl]-1-(5-methylpyrimidin-2-yl)-1-oxidanyl-propane-2-sulfonamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yue, Y, Liu, L.E, Wu, L.J, Xu, F, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7K7K
 
 | |
1JZK
 
 | | Crystal Structure of Scapharca inaequivalvis HbI, I114F mutant (deoxy) | | Descriptor: | GLOBIN I - ARK SHELL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Gibson, Q.H, Cushing, L, Royer Jr, W.E. | | Deposit date: | 2001-09-16 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Restricting the Ligand-Linked Heme Movement in Scapharca Dimeric Hemoglobin Reveals Tight Coupling between Distal and Proximal
Contributions to Cooperativity.
Biochemistry, 40, 2001
|
|
7W0M
 
 | | Cryo-EM structure of a monomeric GPCR-Gi complex with small molecule | | Descriptor: | (1R,2S)-N-[4-(2,6-dimethoxyphenyl)-5-(6-methylpyridin-2-yl)-1,2,4-triazol-3-yl]-1-(5-methylpyrimidin-2-yl)-1-oxidanyl-propane-2-sulfonamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Yue, Y, Liu, L.E, Wu, L.J, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1MCP
 
 | | PHOSPHOCHOLINE BINDING IMMUNOGLOBULIN FAB MC/PC603. AN X-RAY DIFFRACTION STUDY AT 2.7 ANGSTROMS | | Descriptor: | IGA-KAPPA MCPC603 FAB (HEAVY CHAIN), IGA-KAPPA MCPC603 FAB (LIGHT CHAIN), SULFATE ION | | Authors: | Satow, Y, Cohen, G.H, Padlan, E.A, Davies, D.R. | | Deposit date: | 1984-07-09 | | Release date: | 1985-01-02 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Phosphocholine binding immunoglobulin Fab McPC603. An X-ray diffraction study at 2.7 A.
J.Mol.Biol., 190, 1986
|
|
7W0O
 
 | | Cryo-EM structure of a monomeric GPCR-Gi complex with peptide | | Descriptor: | Apelin receptor early endogenous ligand, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Yue, Y, Liu, L.E, Wu, L.J, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
