7X21
 
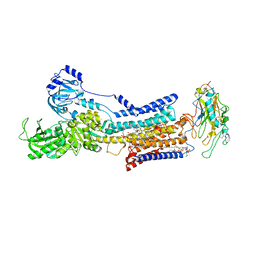 | | Cryo-EM structure of non gastric H,K-ATPase alpha2 K794A in (K+)E2-AlF state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Nakanishi, H, Abe, K. | | Deposit date: | 2022-02-25 | | Release date: | 2022-10-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and function of H + /K + pump mutants reveal Na + /K + pump mechanisms.
Nat Commun, 13, 2022
|
|
7X24
 
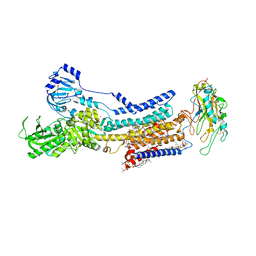 | | Cryo-EM structure of non gastric H,K-ATPase alpha2 SPWC mutant in (2K+)E2-AlF state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Abe, K, Nakanishi, H, Young, V, Artigas, P. | | Deposit date: | 2022-02-25 | | Release date: | 2022-10-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of H + /K + pump mutants reveal Na + /K + pump mechanisms.
Nat Commun, 13, 2022
|
|
7WTO
 
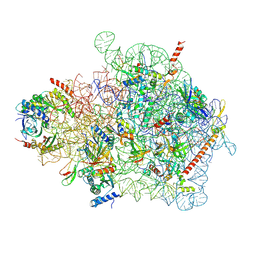 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Tsr1-1 (without Rps2) | | Descriptor: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTN
 
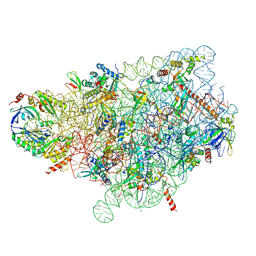 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Tsr1-1 (with Rps2) | | Descriptor: | 18S rRNA, 18S rRNA (guanine(1575)-N(7))-methyltransferase, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTM
 
 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Dis-E | | Descriptor: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Cheng, J, La Venuta, G, Lau, B, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | In vitro structural maturation of an early stage pre-40S particle coupled with U3 snoRNA release and central pseudoknot formation.
Nucleic Acids Res., 50, 2022
|
|
7WTL
 
 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Dis-D | | Descriptor: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Cheng, J, La Venuta, G, Lau, B, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In vitro structural maturation of an early stage pre-40S particle coupled with U3 snoRNA release and central pseudoknot formation.
Nucleic Acids Res., 50, 2022
|
|
7WTR
 
 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Tsr1-3 | | Descriptor: | 18S rRNA, 18S rRNA (guanine(1575)-N(7))-methyltransferase, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTP
 
 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Tsr1-2 (with Rps2) | | Descriptor: | 18S rRNA, 18S rRNA (guanine(1575)-N(7))-methyltransferase, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTT
 
 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-A1 (with CK1) | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTU
 
 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-A1 (without CK1) | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7V7Z
 
 | | Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form
To Be Published
|
|
7V86
 
 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form
To Be Published
|
|
7V7J
 
 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 3
To Be Published
|
|
7V7E
 
 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), one RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), one RBD-up conformation 1
To Be Published
|
|
7V7H
 
 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 1
To Be Published
|
|
7V80
 
 | | Local refinement of SARS-CoV-2 S-Beta variant (B.1.351) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Local refinement of SARS-CoV-2 S-Beta variant (B.1.351) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain
To Be Published
|
|
7V7F
 
 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), one RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), one RBD-up conformation 2
To Be Published
|
|
7V7G
 
 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), two RBD-up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), two RBD-up conformation
To Be Published
|
|
7V87
 
 | | Local refinement of SARS-CoV-2 S-Kappa variant (B.1.617.1) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Angiotensin-converting enzyme 2 (ACE2) ectodomain, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Local refinement of SARS-CoV-2 S-Kappa variant (B.1.617.1) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain
To Be Published
|
|
7WST
 
 | | Cryo-EM structure of the barley Yellow stripe 1 transporter in complex with Fe(III)-DMA | | Descriptor: | (2~{S})-1-[(3~{S})-3-[[(3~{S})-3,4-bis(oxidanyl)-4-oxidanylidene-butyl]amino]-4-oxidanyl-4-oxidanylidene-butyl]azetidine-2-carboxylic acid, CHOLESTEROL HEMISUCCINATE, FE (III) ION, ... | | Authors: | Yamagata, A. | | Deposit date: | 2022-02-01 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Uptake mechanism of iron-phytosiderophore from the soil based on the structure of yellow stripe transporter.
Nat Commun, 13, 2022
|
|
7WSU
 
 | | Cryo-EM structure of the barley Yellow stripe 1 transporter in complex with Fe(III)-PDMA | | Descriptor: | (2~{S})-1-[(3~{S})-3-[[(3~{S})-3,4-bis(oxidanyl)-4-oxidanylidene-butyl]amino]-4-oxidanyl-4-oxidanylidene-butyl]pyrrolidine-2-carboxylic acid, CHOLESTEROL HEMISUCCINATE, FE (III) ION, ... | | Authors: | Yamagata, A. | | Deposit date: | 2022-02-01 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Uptake mechanism of iron-phytosiderophore from the soil based on the structure of yellow stripe transporter.
Nat Commun, 13, 2022
|
|
7WSR
 
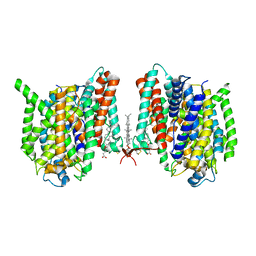 | | Cryo-EM structure of the barley Yellow stripe 1 transporter | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Iron-phytosiderophore transporter | | Authors: | Yamagata, A. | | Deposit date: | 2022-02-01 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Uptake mechanism of iron-phytosiderophore from the soil based on the structure of yellow stripe transporter.
Nat Commun, 13, 2022
|
|
7X91
 
 | |
7X8W
 
 | |
7X92
 
 | |
