8AVY
 
 | | The ABCB1 L335C mutant (mABCB1) in the Apo state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-08-27 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
6SGX
 
 | | Structure of protomer 1 of the ESX-3 core complex | | Descriptor: | ESX-3 secretion system EccB3, ESX-3 secretion system protein EccC3, ESX-3 secretion system protein EccD3, ... | | Authors: | Famelis, N, Rivera-Calzada, A, Llorca, O, Geibel, S. | | Deposit date: | 2019-08-05 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of the mycobacterial type VII secretion system.
Nature, 576, 2019
|
|
6LMR
 
 | | Solution structure of cold shock domain and ssDNA complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*AP*CP*CP*T)-3'), Y-box-binding protein 1 | | Authors: | Fan, J, Yang, D. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA binding to human YB-1 cold shock domain regulated by phosphorylation.
Nucleic Acids Res., 48, 2020
|
|
6EYF
 
 | | Butyrylcolinesterase expressed in CHO cells co-crystallised with a rivastigmine analogue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cholinesterase, GLYCEROL, ... | | Authors: | Brazzolotto, X, De la Mora, E, Dighe, S, Ross, B. | | Deposit date: | 2017-11-12 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rivastigmine and metabolite analogues with putative Alzheimer's disease-modifying properties in a Caenorhabditis elegans model
Commun Chem, 2019
|
|
6LNT
 
 | | Cryo-EM structure of immature Zika virus in complex with human antibody DV62.5 Fab | | Descriptor: | Envelope protein, Fab DV62.5 heavy-chain variable region, Fab DV62.5 light-chain variable region, ... | | Authors: | Tan, T.Y, Fibriansah, G, Kostyuchenko, V.A, Ng, T.S, Lim, X.X, Lim, X.N, Shi, J, Morais, M.C, Corti, D, Lok, S.M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Capsid protein structure in Zika virus reveals the flavivirus assembly process.
Nat Commun, 11, 2020
|
|
6AUT
 
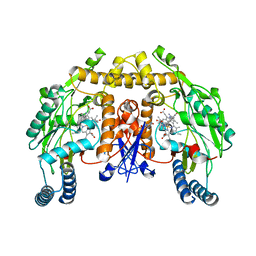 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(3-(3-(Dimethylamino)propyl)-5-fluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{3-[3-(dimethylamino)propyl]-5-fluorophenyl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
3KAD
 
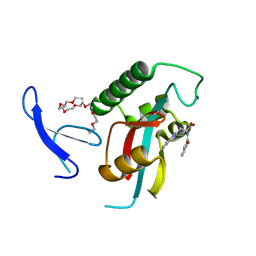 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 3-(1H-benzimidazol-2-yl)-N-(3-phenylpropanoyl)-D-alanine, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8AKF
 
 | |
4E36
 
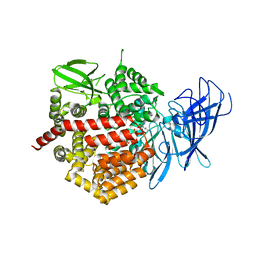 | | Crystal structure of the human Endoplasmic Reticulum Aminopeptidase 2 variant N392K | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Birtley, J.R, Saridakis, E, Pegias, P, Stratikos, E, Mavridis, I.M. | | Deposit date: | 2012-03-09 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A common single nucleotide polymorphism in endoplasmic reticulum aminopeptidase 2 induces a specificity switch that leads to altered antigen processing.
J.Immunol., 189, 2012
|
|
6F12
 
 | | GLIC mutant E181A | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Full mutational mapping of titratable residues helps to identify proton-sensors involved in the control of channel gating in the Gloeobacter violaceus pentameric ligand-gated ion channel.
PLoS Biol., 15, 2017
|
|
6S8H
 
 | | Cryo-EM structure of LptB2FG in complex with LPS | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
6S8N
 
 | | Cryo-EM structure of LptB2FGC in complex with lipopolysaccharide | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, Inner membrane protein yjgQ, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
3J46
 
 | | Structure of the SecY protein translocation channel in action | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L1, 50S ribosomal protein L23P, ... | | Authors: | Akey, C.W, Park, E, Menetret, J.F, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A. | | Deposit date: | 2013-06-18 | | Release date: | 2013-10-23 | | Last modified: | 2019-07-03 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
3J5L
 
 | | Structure of the E. coli 50S subunit with ErmBL nascent chain | | Descriptor: | 23S ribsomal RNA, 5'-R(*CP*(MA6))-3', 5'-R(*CP*CP*A)-3', ... | | Authors: | Arenz, S, Ramu, H, Gupta, P, Berninghausen, O, Beckmann, R, Vazquez-Laslop, N, Mankin, A.S, Wilson, D.N. | | Deposit date: | 2013-10-23 | | Release date: | 2014-03-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide.
Nat Commun, 5, 2014
|
|
6BDL
 
 | |
8ASL
 
 | | RCII/PSI complex, class 2 | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3'R)-3'-hydroxy-beta,beta-caroten-4-one, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhao, Z, Vercellino, I, Knoppova, J, Sobotka, R, Murray, J.W, Nixon, P.J, Sazanov, L.A, Komenda, J. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | The Ycf48 accessory factor occupies the site of the oxygen-evolving manganese cluster during photosystem II biogenesis.
Nat Commun, 14, 2023
|
|
6BE1
 
 | | Cryo-EM structure of serotonin receptor | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Basak, S, Chakrapani, S. | | Deposit date: | 2017-10-24 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Cryo-EM structure of 5-HT
Nat Commun, 9, 2018
|
|
8AKD
 
 | |
8AQY
 
 | | streptavidin mutant S112A with an iridium catalyst for CH activation | | Descriptor: | Streptavidin, tert-butyl 7'-[5-[(3aS,4S,6aR)-2-oxidanylidene-1,3,3a,4,6,6a-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]-1-chloranyl-2,3,4,5,6-pentamethyl-spiro[1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane-1,2'-3-aza-1-azonia-2$l^{8}-iridatricyclo[6.3.1.0^{4,12}]dodeca-1(11),4,6,8(12),9-pentaene]-3'-carboxylate | | Authors: | Igareta, N.V, Ward, T.R. | | Deposit date: | 2022-08-15 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | streptavidin mutant S112A with an iridium catalyst for CH activation
To Be Published
|
|
3EX8
 
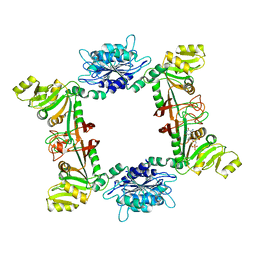 | | Complex structure of bacillus subtilis RibG reduction mechanism in riboflavin biosynthesis | | Descriptor: | Riboflavin biosynthesis protein ribD, ZINC ION, [(2R,3S,4R,5E)-5-[(5-amino-2,6-dioxo-3H-pyrimidin-4-yl)imino]-2,3,4-trihydroxy-pentyl] dihydrogen phosphate | | Authors: | Chen, S.C, Lin, Y.H, Yu, H.C, Liaw, S.H. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Complex structure of Bacillus subtilis RibG: the reduction mechanism during riboflavin biosynthesis.
J.Biol.Chem., 284, 2009
|
|
8AK6
 
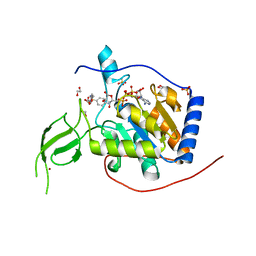 | |
8AK9
 
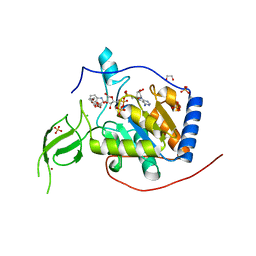 | |
3FG4
 
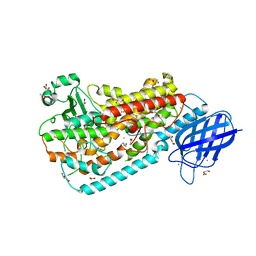 | | Crystal structure of Delta413-417:GS I805A LOX | | Descriptor: | ACETIC ACID, ARACHIDONIC ACID, Allene oxide synthase-lipoxygenase protein, ... | | Authors: | Neau, D.B, Newcomer, M.E. | | Deposit date: | 2008-12-04 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The 1.85 A structure of an 8R-lipoxygenase suggests a general model for lipoxygenase product specificity.
Biochemistry, 48, 2009
|
|
4E0S
 
 | | Crystal Structure of C5b-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C5, ... | | Authors: | Aleshin, A.E, Stec, B, DiScipio, R, Liddington, R.C. | | Deposit date: | 2012-03-05 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.21 Å) | | Cite: | Crystal structure of c5b-6 suggests structural basis for priming assembly of the membrane attack complex.
J.Biol.Chem., 287, 2012
|
|
8AKE
 
 | |
