1JL3
 
 | | Crystal Structure of B. subtilis ArsC | | Descriptor: | ARSENATE REDUCTASE, SULFATE ION | | Authors: | Su, X.-D, Bennett, M.S. | | Deposit date: | 2001-07-15 | | Release date: | 2001-10-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bacillus subtilis arsenate reductase is structurally and functionally similar to low molecular weight protein tyrosine phosphatases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3O1W
 
 | | Crystal structure of dimeric KlHxk1 in crystal form III | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Hexokinase, ... | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-22 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
1JII
 
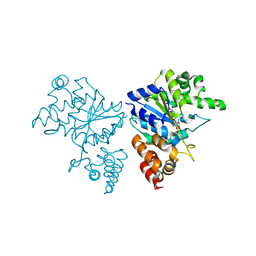 | | Crystal structure of S. aureus TyrRS in complex with SB-219383 | | Descriptor: | [2-AMINO-3-(4-HYDROXY-PHENYL)-PROPIONYLAMINO]- (2,4,5,8-TETRAHYDROXY-7-OXA-2-AZA-BICYCLO[3.2.1]OCT-3-YL)- ACETIC ACID, tyrosyl-tRNA synthetase | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Jarvest, R.L. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors.
Protein Sci., 10, 2001
|
|
3O43
 
 | |
3O1B
 
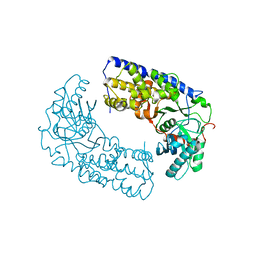 | | CRYSTAL STRUCTURE OF DIMERIC KLHXK1 IN CRYSTAL FORM II | | Descriptor: | Hexokinase | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-21 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O2L
 
 | |
3O40
 
 | | Complex of a chimeric alpha/beta-peptide based on the gp41 CHR domain bound to gp41-5 | | Descriptor: | CHLORIDE ION, GLYCEROL, NONAETHYLENE GLYCOL, ... | | Authors: | Horne, W.S, Johnson, L.M, Gellman, S.H. | | Deposit date: | 2010-07-26 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Broad Distribution of Energetically Important Contacts across an Extended Protein Interface.
J.Am.Chem.Soc., 133, 2011
|
|
3P85
 
 | |
3OOI
 
 | | Crystal Structure of Human Histone-Lysine N-methyltransferase NSD1 SET domain in Complex with S-adenosyl-L-methionine | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific, S-ADENOSYLMETHIONINE, ... | | Authors: | Qiao, Q, Wang, M, Xu, R.M. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation
J.Biol.Chem., 286, 2010
|
|
1IRX
 
 | | Crystal structure of class I lysyl-tRNA synthetase | | Descriptor: | ZINC ION, lysyl-tRNA synthetase | | Authors: | Nureki, O, Terada, T, Ishitani, R, Ambrogelly, A, Ibba, M, Soll, D, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-10-25 | | Release date: | 2002-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional convergence of two lysyl-tRNA synthetases with unrelated topologies.
Nat.Struct.Biol., 9, 2002
|
|
3PD8
 
 | | X-ray structure of the ligand-binding core of GluA2 in complex with (S)-7-HPCA at 2.5 A resolution | | Descriptor: | (7S)-3-hydroxy-4,5,6,7-tetrahydroisoxazolo[5,4-c]pyridine-7-carboxylic acid, ACETIC ACID, CACODYLATE ION, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.476 Å) | | Cite: | Biostructural and pharmacological studies of bicyclic analogues of the 3-isoxazolol glutamate receptor agonist ibotenic acid.
J. Med. Chem., 53, 2010
|
|
3PDY
 
 | |
1L6P
 
 | |
3PE0
 
 | |
3PD9
 
 | | X-ray structure of the ligand-binding core of GluA2 in complex with (R)-5-HPCA at 2.1 A resolution | | Descriptor: | (5R)-3-hydroxy-4,5,6,7-tetrahydroisoxazolo[5,4-c]pyridine-5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biostructural and pharmacological studies of bicyclic analogues of the 3-isoxazolol glutamate receptor agonist ibotenic acid.
J. Med. Chem., 53, 2010
|
|
1ILT
 
 | |
3PEO
 
 | | Crystal structure of acetylcholine binding protein complexed with metocurine | | Descriptor: | 6,6',7',12'-tetramethoxy-2,2,2',2'-tetramethyltubocuraran-2,2'-diium, Soluble acetylcholine receptor | | Authors: | Talley, T.T, Harel, M, Yamauchi, G.J, Radic, Z, Hansen, S, Huxford, T, Taylor, P.W. | | Deposit date: | 2010-10-27 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The curare alkaloids: analyzing the poses of complexes with the acetylcholine binding protein in relation to structure and binding energetics
To be Published
|
|
3PLW
 
 | | Ref protein from P1 bacteriophage | | Descriptor: | Recombination enhancement function protein, SULFATE ION, ZINC ION | | Authors: | Keck, J.L, Lu, D, Cox, M.M. | | Deposit date: | 2010-11-15 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Creating Directed Double-strand Breaks with the Ref Protein: A NOVEL RecA-DEPENDENT NUCLEASE FROM BACTERIOPHAGE P1.
J.Biol.Chem., 286, 2011
|
|
3PMT
 
 | | Crystal structure of the Tudor domain of human Tudor domain-containing protein 3 | | Descriptor: | TETRAETHYLENE GLYCOL, Tudor domain-containing protein 3 | | Authors: | Lam, R, Bian, C.B, Guo, Y.H, Xu, C, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of TDRD3 and Methyl-Arginine Binding Characterization of TDRD3, SMN and SPF30.
Plos One, 7, 2012
|
|
3PMZ
 
 | | Crystal Structure of the Complex of Acetylcholine Binding Protein and d-tubocurarine | | Descriptor: | (1beta,1'alpha)-7',12'-dihydroxy-6,6'-dimethoxy-2,2',2'-trimethyltubocuraran-2'-ium, MAGNESIUM ION, Soluble acetylcholine receptor | | Authors: | Talley, T.T, Harel, M, Yamauchi, J.G, Radic, Z, Hansen, S, Huxford, T, Taylor, P.W. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The Curare Alkaloids: Analyzing the Poses of Complexes with the Acetylcholine Binding Protein in Relation to Structure and Binding Energies
To be Published
|
|
3OJ6
 
 | |
3OHT
 
 | | Crystal Structure of Salmo Salar p38alpha | | Descriptor: | N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, SULFATE ION, p38a | | Authors: | Rothweiler, U, Johnson, K, Engh, R.A. | | Deposit date: | 2010-08-18 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | p38alpha MAP kinase dimers with swapped activation segments and a novel catalytic loop conformation
J.Mol.Biol., 411, 2011
|
|
3OWD
 
 | | Crystal Structure of HSP90 with N-Aryl-benzimidazolone II | | Descriptor: | Heat shock protein HSP 90-alpha, N-{[1-(5-chloro-2,4-dihydroxyphenyl)-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl]methyl}naphthalene-1-sulfonamide | | Authors: | Park, C.H. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | N-aryl-benzimidazolones as novel small molecule HSP90 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3PJN
 
 | | The crystal structure of Tp34 bound to Zn(II) ion at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, SULFATE ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Machius, M, Norgard, M.V. | | Deposit date: | 2010-11-10 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of the Tp34 lipoprotein from Treponema pallidum suggests a role in transition metal homeostasis
To be Published
|
|
1IRO
 
 | | RUBREDOXIN (OXIDIZED, FE(III)) AT 1.1 ANGSTROMS RESOLUTION | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Dauter, Z, Wilson, K.S, Sieker, L.C, Moulis, J.M, Meyer, J. | | Deposit date: | 1995-12-13 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Zinc- and iron-rubredoxins from Clostridium pasteurianum at atomic resolution: a high-precision model of a ZnS4 coordination unit in a protein.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
