8CTI
 
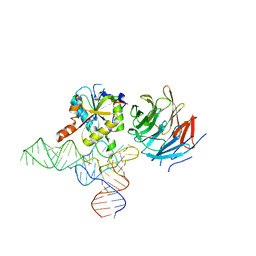 | | Cryo-EM structure of human METTL1-WDR4-tRNA(Val) complex | | Descriptor: | tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4, tRNA-Val-TAC-2-1 | | Authors: | Li, J, Wang, L, Fontana, P, Hunkeler, M, Roy-Burman, S.S, Wu, H, Fischer, E.S, Gregory, R.I. | | Deposit date: | 2022-05-14 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
5AJL
 
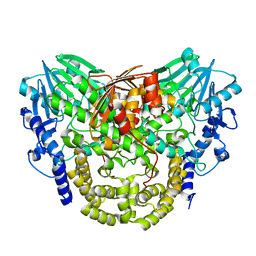 | | Sdsa sulfatase tetragonal | | Descriptor: | ALKYL SULFATASE, ZINC ION | | Authors: | De la Mora, E, Flores-Hernandez, E, Jakoncic, J, Stojanoff, V, Sanchez-Puig, N, Moreno, A. | | Deposit date: | 2015-02-25 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Sdsa Polymorph Isolation and Improvement of Their Crystal Quality Using Nonconventional Crystallization Techniques
J.Appl.Crystallogr., 48, 2015
|
|
6B7B
 
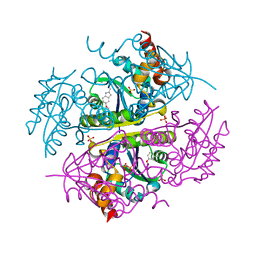 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 5-methoxy-2-methyl-1H-indole | | Descriptor: | 5-methoxy-2-methyl-1H-indole, DIMETHYL SULFOXIDE, PYROPHOSPHATE 2-, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
5IS0
 
 | | Structure of TRPV1 in complex with capsazepine, determined in lipid nanodisc | | Descriptor: | Transient receptor potential cation channel subfamily V member 1, capsazepine | | Authors: | Gao, Y, Cao, E, Julius, D, Cheng, Y. | | Deposit date: | 2016-03-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action.
Nature, 534, 2016
|
|
6JKT
 
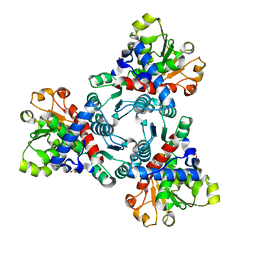 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with N-(PHOSPHONACETYL)-L-ASPARTIC ACID (PALA). | | Descriptor: | Aspartate carbamoyltransferase, GLYCEROL, N-(PHOSPHONACETYL)-L-ASPARTIC ACID | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
2OWS
 
 | |
2BS4
 
 | | GLU C180 -> ILE VARIANT QUINOL:FUMARATE REDUCTASE FROMWOLINELLA SUCCINOGENES | | Descriptor: | 2,3-DIMETHYL-1,4-NAPHTHOQUINONE, CITRIC ACID, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Lancaster, C.R.D. | | Deposit date: | 2005-05-14 | | Release date: | 2005-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Experimental Support for the E-Pathway Hypothesis of Coupled Transmembrane Electron and Proton Transfer in Dihemic Quinol:Fumarate Reductase
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6O9A
 
 | | Crystal structure of MqnA complexed with 3-hydroxybenzoic acid | | Descriptor: | 3-HYDROXYBENZOIC ACID, ACETATE ION, Chorismate dehydratase | | Authors: | Hicks, K.A, Mahanta, N, Naseem, S, Fedoseyenko, D, Begley, T.P, Ealick, S.E. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.326 Å) | | Cite: | Menaquinone Biosynthesis: Biochemical and Structural Studies of Chorismate Dehydratase.
Biochemistry, 58, 2019
|
|
8D47
 
 | |
1YRS
 
 | | Crystal structure of KSP in complex with inhibitor 1 | | Descriptor: | 3-[(5S)-1-ACETYL-3-(2-CHLOROPHENYL)-4,5-DIHYDRO-1H-PYRAZOL-5-YL]PHENOL, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Cox, C.D, Breslin, M.J, Mariano, B.J, Coleman, P.J, Buser, C.A, Walsh, E.S, Hamilton, K, Kohl, N.E, Torrent, M, Yan, Y, Kuo, L.C, Hartman, G.D. | | Deposit date: | 2005-02-04 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 1: The discovery of 3,5-diaryl-4,5-dihydropyrazoles as potent and selective inhibitors of the mitotic kinesin KSP
BIOORG.MED.CHEM.LETT., 15, 2005
|
|
4N1B
 
 | | STRUCTURE OF KEAP1 KELCH DOMAIN WITH(1S,2R)-2-[(1S)-1-[(1-oxo-2,3-dihydro-1H-isoindol-2-Yl)methyl]-1,2,3,4-tetrahydroisoquinoline-2-Carbonyl]cyclohexane-1-carboxylic acid | | Descriptor: | (1S,2R)-2-{[(1S)-1-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1 | | Authors: | Smith, M.A, Duclos, S, Beaumont, E, Kwong, J, Brooks, M, Barker, J, Jnoff, E, Brookfield, F, Courade, J.P, Barker, O, Fryatt, T, Albrecht, C, Bromidge, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-02-19 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Binding Mode and Structure-Activity Relationships around Direct Inhibitors of the Nrf2-Keap1 Complex.
Chemmedchem, 9, 2014
|
|
6Q8E
 
 | | Crystal structure of branched-chain amino acid aminotransferase from Thermobaculum terrenum in PMP-form | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Branched-chain-amino-acid aminotransferase, CHLORIDE ION | | Authors: | Boyko, K.M, Bezsudnova, E.Y, Nikolaeva, A.Y, Zeifman, Y.S, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2018-12-14 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and structural insights into PLP fold type IV transaminase from Thermobaculum terrenum.
Biochimie, 158, 2018
|
|
3OIC
 
 | | Crystal Structure of Enoyl-ACP Reductases III (FabL) from B. subtilis (apo form) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH], SULFATE ION | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
1I79
 
 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COVALENTLY BOUND 5'-DEOXY-5'-[(3-HYDRAZINOPROPYL)METHYLAMINO]ADENOSINE | | Descriptor: | 1,4-DIAMINOBUTANE, 5'-DEOXY-5'-[(3-HYDRAZINOPROPYL)METHYLAMINO]ADENOSINE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-08 | | Release date: | 2001-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
7BV2
 
 | | The nsp12-nsp7-nsp8 complex bound to the template-primer RNA and triphosphate form of Remdesivir(RTP) | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
7BV1
 
 | | Cryo-EM structure of the apo nsp12-nsp7-nsp8 complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
3OK0
 
 | | E35A Mutant of Hen Egg White Lysozyme (HEWL) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | O'Meara, F, Bradley, J, O'Rourke, P.E, Webb, H, Tynan-Connolly, B.M, Nielsen, J.E. | | Deposit date: | 2010-08-24 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | E35A Mutant of Hen Egg White Lysozyme (HEWL)
TO BE PUBLISHED
|
|
8TR5
 
 | | Cryo-EM structure of the rat P2X7 receptor in the apo closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GUANOSINE-5'-DIPHOSPHATE, P2X purinoceptor 7, ... | | Authors: | Oken, A.C, Lisi, N.E, Krishnamurthy, I, McCarthy, A.E, Godsey, M.H, Glasfeld, A, Mansoor, S.E. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | High-affinity agonism at the P2X7 receptor is mediated by three residues outside the orthosteric pocket
Nat Commun, 2024
|
|
8TRJ
 
 | | Cryo-EM structure of the rat P2X7 receptor in complex with the high-affinity agonist BzATP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3'-O-(4-benzoylbenzoyl)adenosine 5'-(tetrahydrogen triphosphate), GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Oken, A.C, Lisi, N.E, Krishnamurthy, I, McCarthy, A.E, Godsey, M.H, Glasfeld, A, Mansoor, S.E. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | High-affinity agonism at the P2X7 receptor is mediated by three residues outside the orthosteric pocket
Nat Commun, 2024
|
|
1I2L
 
 | | DEOXYCHORISMATE LYASE FROM ESCHERICHIA COLI WITH INHIBITOR | | Descriptor: | 4-AMINO-4-DEOXYCHORISMATE LYASE, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Jensen, P.Y, Parsons, J.F, Fisher, K.E, Pachikara, A.S, Tordova, M, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-02-09 | | Release date: | 2003-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of Escherichia coli Aminodeoxychorismate Lyase
To be Published
|
|
1I2W
 
 | | BETA-LACTAMASE FROM BACILLUS LICHENIFORMIS BS3 COMPLEXED WITH CEFOXITIN | | Descriptor: | (2R)-5-[(carbamoyloxy)methyl]-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, BETA-LACTAMASE, CARBAMIC ACID | | Authors: | Fonze, E, Vanhove, M, Dive, G, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-02-12 | | Release date: | 2002-03-13 | | Last modified: | 2018-09-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the Bacillus licheniformis BS3 class A beta-lactamase and of the acyl-enzyme adduct formed with cefoxitin
Biochemistry, 41, 2002
|
|
6BZH
 
 | | Structure of mouse RIG-I tandem CARDs | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Probable ATP-dependent RNA helicase DDX58 | | Authors: | Kershaw, N.J, D'Cruz, A.A, Babon, J.J, Nicholson, S.E. | | Deposit date: | 2017-12-24 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a second binding site on the TRIM25 B30.2 domain.
Biochem. J., 475, 2018
|
|
8V4S
 
 | | Cryo-EM structure of the rat P2X7 receptor in the apo closed state purified in the absence of sodium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GUANOSINE-5'-DIPHOSPHATE, P2X purinoceptor 7, ... | | Authors: | Oken, A.C, Lisi, N.E, Krishnamurthy, I, McCarthy, A.E, Godsey, M.H, Glasfeld, A, Mansoor, S.E. | | Deposit date: | 2023-11-29 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | High-affinity agonism at the P2X7 receptor is mediated by three residues outside the orthosteric pocket
Nat Commun, 2024
|
|
3OJP
 
 | | D52N Mutant of Hen Egg White Lysozyme (HEWL) | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | O'Meara, F, Bradley, J, O'Rourke, P.E, Webb, H, Tynan-Connolly, B.M, Nielsen, J.E. | | Deposit date: | 2010-08-23 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | D52N Mutant of Hen Egg White Lysozyme (HEWL)
TO BE PUBLISHED
|
|
2VTJ
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design | | Descriptor: | 4-[(6-chloropyrazin-2-yl)amino]benzenesulfonamide, CELL DIVISION PROTEIN KINASE 2, GLYCEROL | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
