7C88
 
 | | Complex structure of JS003 and PD-L1 | | Descriptor: | JS003 Heavy chain, JS003 Light chain, Programmed cell death 1 ligand 1 | | Authors: | Bi, X, Shi, R, Chai, Y, Qi, J, Yan, J, Tan, S. | | Deposit date: | 2020-05-29 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Identification of a hotspot on PD-L1 for pH-dependent binding by monoclonal antibodies for tumor therapy.
Signal Transduct Target Ther, 5, 2020
|
|
5IF3
 
 | |
7AUW
 
 | | Inhibitory complex of human meprin beta with mouse fetuin-B. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fetuin-B, ... | | Authors: | Eckhard, U, Gomis-Ruth, F.X. | | Deposit date: | 2020-11-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of a 250-kDa heterotetrameric particle explains inhibition of sheddase meprin beta by endogenous fetuin-B.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6KA7
 
 | | The complex structure of Human IgG Fc and its binding Repebody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin gamma-1 heavy chain, repebody | | Authors: | Choi, J, Kim, H. | | Deposit date: | 2019-06-21 | | Release date: | 2020-06-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The complex structure of Human IgG Fc and its binding Repebody
To Be Published
|
|
5KOE
 
 | | The structure of Arabidopsis thaliana FUT1 in complex with XXLG | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2016-06-30 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural, mutagenic and in silico studies of xyloglucan fucosylation in Arabidopsis thaliana suggest a water-mediated mechanism.
Plant J., 91, 2017
|
|
5L18
 
 | |
5L17
 
 | |
5KKB
 
 | | Structure of mouse Golgi alpha-1,2-mannosidase IA and Man9GlcNAc2-PA complex | | Descriptor: | 1,4-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ... | | Authors: | Xiang, Y, Moremen, K.W. | | Deposit date: | 2016-06-21 | | Release date: | 2016-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | Substrate recognition and catalysis by GH47 alpha-mannosidases involved in Asn-linked glycan maturation in the mammalian secretory pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6HG0
 
 | | Influenza A Virus N9 Neuraminidase complex with NANA (Tern/Australia). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
5L0E
 
 | | Crystal Structure of Autotaxin and Compound 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(3-{2-[(2,3-dihydro-1H-inden-2-yl)amino]-7,8-dihydropyrido[4,3-d]pyrimidin-6(5H)-yl}propanoyl)-1,3-benzoxazol-2(3H)-one, CHLORIDE ION, ... | | Authors: | Durbin, J.D. | | Deposit date: | 2016-07-27 | | Release date: | 2016-08-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Novel Autotaxin Inhibitors for the Treatment of Osteoarthritis Pain: Lead Optimization via Structure-Based Drug Design.
Acs Med.Chem.Lett., 7, 2016
|
|
6I02
 
 | | Structure of human D-glucuronyl C5 epimerase in complex with product | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debarnot, C, Monneau, Y.R, Roig-Zamboni, V, Le Narvor, C, Goulet, A, Fadel, F, Vives, R.R, Bonnaffe, D, Lortat-Jacob, H, Bourne, Y. | | Deposit date: | 2018-10-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1TB6
 
 | | 2.5A Crystal Structure of the Antithrombin-Thrombin-Heparin Ternary Complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfonato-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-alpha-L-idopyranuronic acid-(1-4)-methyl 3-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Johnson, D.J, Esmon, C.T, Huntington, J.A. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the antithrombin-thrombin-heparin ternary complex reveals the antithrombotic mechanism of heparin.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T4X
 
 | | The first left-handed RNA structure of (CGCGCG)2, Z-RNA, NMR, 12 structures, determined in high salt | | Descriptor: | RNA (5'-R(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Popenda, M, Milecki, J, Adamiak, R.W. | | Deposit date: | 2004-04-30 | | Release date: | 2004-08-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High salt solution structure of a left-handed RNA double helix.
Nucleic Acids Res., 32, 2004
|
|
6JAX
 
 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with chitooctaose [(GlcN)8] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
7D3E
 
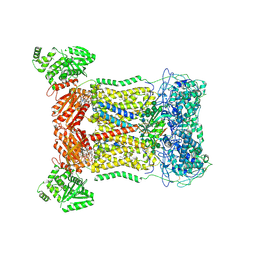 | | Cryo-EM structure of human DUOX1-DUOXA1 in low-calcium state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dual oxidase 1, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chen, L, Wu, J.X. | | Deposit date: | 2020-09-19 | | Release date: | 2020-12-09 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of human dual oxidase 1 complex in low-calcium and high-calcium states.
Nat Commun, 12, 2021
|
|
1TYW
 
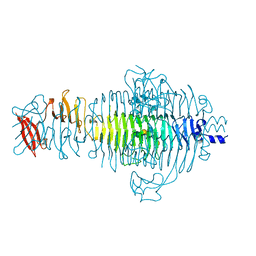 | | STRUCTURE OF TAILSPIKE-PROTEIN | | Descriptor: | TAILSPIKE PROTEIN, alpha-D-galactopyranose-(1-2)-[alpha-D-Tyvelopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-alpha-L-rhamnopyranose-(1-3)-[alpha-D-glucopyranose-(1-4)]alpha-D-galactopyranose-(1-2)-[alpha-D-Tyvelopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-alpha-L-rhamnopyranose | | Authors: | Steinbacher, S, Huber, R. | | Deposit date: | 1996-07-26 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
6I1C
 
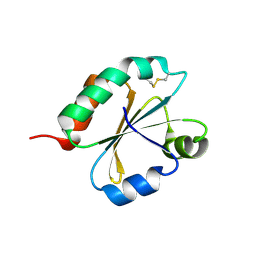 | | Crystal structure of Chlamydomonas reinhardtii thioredoxin f2 | | Descriptor: | thioredoxin f2 | | Authors: | Lemaire, S.D, Tedesco, D, Crozet, P, Michelet, L, Fermani, S, Zaffagnini, M, Henri, J. | | Deposit date: | 2018-10-28 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Chloroplastic Thioredoxin f2 fromChlamydomonas reinhardtiiReveals Distinct Surface Properties.
Antioxidants (Basel), 7, 2018
|
|
6KRV
 
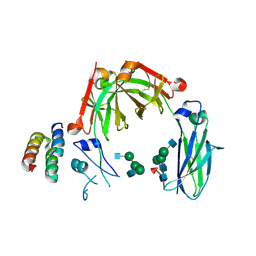 | | Crystal structure of mouse IgG2b Fc complexed with B domain of Protein A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-2B chain C region, ... | | Authors: | Taniguchi, Y, Satoh, T, Yagi, H, Kato, K. | | Deposit date: | 2019-08-22 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | On-Membrane Dynamic Interplay between Anti-GM1 IgG Antibodies and Complement Component C1q.
Int J Mol Sci, 21, 2019
|
|
1SG4
 
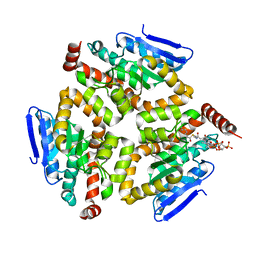 | | Crystal structure of human mitochondrial delta3-delta2-enoyl-CoA isomerase | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, mitochondrial, OCTANOYL-COENZYME A | | Authors: | Partanen, S.T, Novikov, D.K, Popov, A.N, Mursula, A.M, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2004-02-23 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A crystal structure of human mitochondrial Delta3-Delta2-enoyl-CoA isomerase shows a novel mode of binding for the fatty acyl group.
J.Mol.Biol., 342, 2004
|
|
1SLB
 
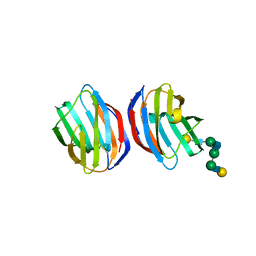 | | X-RAY CRYSTALLOGRAPHY REVEALS CROSSLINKING OF MAMMALIAN LECTIN (GALECTIN-1) BY BIANTENNARY COMPLEX TYPE SACCHARIDES | | Descriptor: | BOVINE GALECTIN-1, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bourne, Y, Cambillau, C. | | Deposit date: | 1994-03-12 | | Release date: | 1995-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crosslinking of mammalian lectin (galectin-1) by complex biantennary saccharides.
Nat.Struct.Biol., 1, 1994
|
|
1W7L
 
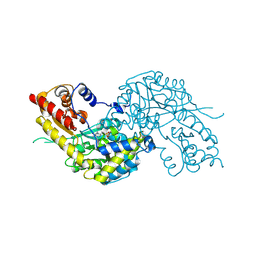 | | Crystal structure of human kynurenine aminotransferase I | | Descriptor: | KYNURENINE--OXOGLUTARATE TRANSAMINASE I, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Rossi, F, Han, Q, Li, J, Li, J, Rizzi, M. | | Deposit date: | 2004-09-06 | | Release date: | 2004-09-08 | | Last modified: | 2015-12-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Kynurenine Aminotransferase I
J.Biol.Chem., 279, 2004
|
|
1VFU
 
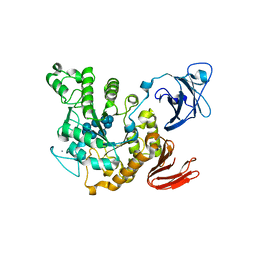 | | Crystal structure of Thermoactinomyces vulgaris R-47 amylase 2/gamma-cyclodextrin complex | | Descriptor: | CALCIUM ION, Cyclooctakis-(1-4)-(alpha-D-glucopyranose), Neopullulanase 2 | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2004-04-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
5A3P
 
 | | Crystal structure of the catalytic domain of human PLU1 (JARID1B). | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Nowak, R, Srikannathasan, V, Johansson, C, Gileadi, C, Tallant, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-06-02 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5A3W
 
 | | Crystal structure of human PLU-1 (JARID1B) in complex with Pyridine-2, 6-dicarboxylic Acid (PDCA) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Kopec, J, Strain-Damerell, C, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-06-03 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
6V8Z
 
 | |
