2G6Y
 
 | |
2G83
 
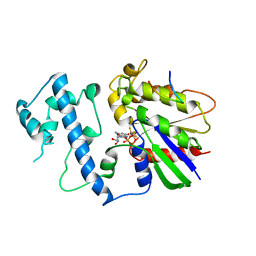 | | Structure of activated G-alpha-i1 bound to a nucleotide-state-selective peptide: Minimal determinants for recognizing the active form of a G protein alpha subunit | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Johnston, C.A, Ramer, J.K, Blaesius, R, Kuhlman, B, Arshavsky, V.Y, Siderovski, D.P. | | Deposit date: | 2006-03-01 | | Release date: | 2006-10-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Minimal Determinants for Binding Activated Galpha from the Structure of a Galpha(i1)-Peptide Dimer.
Biochemistry, 45, 2006
|
|
2QTQ
 
 | |
2QE6
 
 | |
2G1U
 
 | |
2GDN
 
 | |
2GB4
 
 | |
2QEZ
 
 | |
2GBT
 
 | | C6A/C111A CuZn Superoxide dismutase | | Descriptor: | COPPER (I) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Hornberg, A, Logan, D.T, Marklund, S.L, Oliveberg, M. | | Deposit date: | 2006-03-11 | | Release date: | 2007-01-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Coupling between Disulphide Status, Metallation and Dimer Interface Strength in Cu/Zn Superoxide Dismutase
J.Mol.Biol., 365, 2007
|
|
2QYV
 
 | |
2GGU
 
 | |
2GFH
 
 | |
2GIC
 
 | | Crystal Structure of a vesicular stomatitis virus nucleocapsid-RNA complex | | Descriptor: | 45-MER, Nucleocapsid protein, URANYL (VI) ION | | Authors: | Green, T.J, Zhang, X, Wertz, G.W, Luo, M. | | Deposit date: | 2006-03-28 | | Release date: | 2006-08-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structure of the vesicular stomatitis virus nucleoprotein-RNA complex unveils how the RNA is sequestered
Science, 313, 2006
|
|
5JZY
 
 | | Thrombin in complex with (S)-1-((R)-2-amino-3-cyclohexylpropanoyl)-N-(4-carbamimidoylbenzyl)pyrrolidine-2-carboxamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-cyclohexyl-D-alanyl-N-[(4-carbamimidoylphenyl)methyl]-L-prolinamide, DIMETHYL SULFOXIDE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G. | | Deposit date: | 2016-05-17 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Strategies for Late-Stage Optimization: Profiling Thermodynamics by Preorganization and Salt Bridge Shielding.
J.Med.Chem., 62, 2019
|
|
2R2O
 
 | | Crystal structure of the effector domain of human Plexin B1 | | Descriptor: | Plexin-B1, UNKNOWN ATOM OR ION | | Authors: | Tong, Y, Tempel, W, Shen, L, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-27 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of Rac1, Rnd1, and RhoD to a novel Rho GTPase interaction motif destabilizes dimerization of the plexin-B1 effector domain.
J.Biol.Chem., 282, 2007
|
|
2GG5
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | 5-IMINO-4-(2-TRIFLUOROMETHYL-PHENYLAZO)-5H-PYRAZOL-3-YLAMINE, COBALT (II) ION, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
2GHS
 
 | |
5JYU
 
 | |
2QUQ
 
 | |
2G02
 
 | | Nisin cyclase | | Descriptor: | Nisin biosynthesis protein nisC, ZINC ION | | Authors: | Nair, S.K. | | Deposit date: | 2006-02-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanism of the Lantibiotic Cyclase Involved in Nisin Biosynthesis
Science, 311, 2006
|
|
2QCV
 
 | |
2QW9
 
 | | Crystal structure of bovine hsc70 (1-394aa)in the apo state | | Descriptor: | GLYCEROL, Heat shock cognate 71 kDa protein | | Authors: | Jiang, J, Maes, E.G, Wang, L, Taylor, A.B, Hinck, A.P, Lafer, E.M, Sousa, R. | | Deposit date: | 2007-08-10 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of J cochaperone binding and regulation of Hsp70.
Mol.Cell, 28, 2007
|
|
2G37
 
 | | Structure of Thermus thermophilus L-proline dehydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, proline dehydrogenase/delta-1-pyrroline-5-carboxylate dehydrogenase | | Authors: | Tanner, J.J, White, T.A. | | Deposit date: | 2006-02-17 | | Release date: | 2007-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Kinetics of Monofunctional Proline Dehydrogenase from Thermus thermophilus.
J.Biol.Chem., 282, 2007
|
|
2QXI
 
 | | High resolution structure of Human Kallikrein 7 in Complex with Suc-Ala-Ala-Pro-Phe-chloromethylketone | | Descriptor: | Kallikrein-7, N-(3-carboxypropanoyl)-L-alanyl-L-alanyl-N-[(2S,3S)-4-chloro-3-hydroxy-1-phenylbutan-2-yl]-L-prolinamide | | Authors: | Debela, M, Hess, P, Magdolen, V, Schechter, N.M, Bode, W, Goettig, P. | | Deposit date: | 2007-08-11 | | Release date: | 2008-01-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Chymotryptic specificity determinants in the 1.0 A structure of the zinc-inhibited human tissue kallikrein 7.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2G7B
 
 | |
