5ET3
 
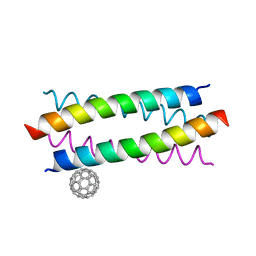 | | Crystal Structure of De novo Designed Fullerene organizing peptide | | Descriptor: | (C_{60}-I_{h})[5,6]fullerene, Fullerene Organizing Protein (C60Sol-COP-3) | | Authors: | Kim, K.-H, Kim, Y.H, Acharya, R, Kim, N.H, Paul, J, Grigoryan, G, DeGrado, W.F. | | Deposit date: | 2015-11-17 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.671 Å) | | Cite: | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
6TBO
 
 | |
6TBP
 
 | |
6T71
 
 | |
6TAW
 
 | |
6T3G
 
 | |
6TAL
 
 | | 3C-like protease from Southampton virus complexed with FMOPL000227a. | | Descriptor: | 5-ethyl-1,3,4-thiadiazol-2-amine, DIMETHYL SULFOXIDE, Genome polyprotein, ... | | Authors: | Guo, J, Cooper, J.B. | | Deposit date: | 2019-10-29 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | In crystallo-screening for discovery of human norovirus 3C-like protease inhibitors.
J Struct Biol X, 4, 2020
|
|
1FWS
 
 | | AQUIFEX AEOLICUS KDO8P SYNTHASE IN COMPLEX WITH PEP AND CADMIUM | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, CADMIUM ION, PHOSPHATE ION, ... | | Authors: | Duewel, H.S, Radaev, S, Wang, J, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2000-09-24 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Electronic structure of the metal center in the Cd(2+), Zn(2+), and Cu(2+) substituted forms of KDO8P synthase: implications for catalysis.
Biochemistry, 48, 2009
|
|
1YRR
 
 | | Crystal Structure Of The N-Acetylglucosamine-6-Phosphate Deacetylase From Escherichia Coli K12 at 2.0 A Resolution | | Descriptor: | GLYCEROL, N-acetylglucosamine-6-phosphate deacetylase, PHOSPHATE ION | | Authors: | Ferreira, F.M, Aparicio, R, Mendoza-Hernandez, G, Calcagno, M.L, Oliva, G. | | Deposit date: | 2005-02-04 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of N-acetylglucosamine-6-phosphate deacetylase apoenzyme from Escherichia coli.
J.Mol.Biol., 359, 2006
|
|
3R9Y
 
 | | Crystal Structure of StWhy2 K67A (form I) | | Descriptor: | PHOSPHATE ION, Why2 protein | | Authors: | Cappadocia, L, Brisson, N, Sygusch, J. | | Deposit date: | 2011-03-26 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A conserved lysine residue of plant Whirly proteins is necessary for higher order protein assembly and protection against DNA damage.
Nucleic Acids Res., 40, 2012
|
|
1FWW
 
 | | AQUIFEX AEOLICUS KDO8P SYNTHASE IN COMPLEX WITH PEP, A5P AND CADMIUM | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, ARABINOSE-5-PHOSPHATE, CADMIUM ION, ... | | Authors: | Duewel, H.S, Radaev, S, Wang, J, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2000-09-24 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Electronic structure of the metal center in the Cd(2+), Zn(2+), and Cu(2+) substituted forms of KDO8P synthase: implications for catalysis.
Biochemistry, 48, 2009
|
|
1RJW
 
 | |
4BZW
 
 | | Complete crystal structure of the carboxylesterase Cest-2923 (lp_2923) from Lactobacillus plantarum WCFS1 | | Descriptor: | ACETATE ION, LIPASE/ESTERASE, SULFATE ION, ... | | Authors: | Benavente, R, Esteban-Torres, M, Acebron, I, delasRivas, B, Munoz, R, Alvarez, Y, Mancheno, J.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Structure, Biochemical Characterization and Analysis of the Pleomorphism of Carboxylesterase Cest-2923 from Lactobacillus Plantarum Wcfs1
FEBS J., 280, 2013
|
|
6TCF
 
 | |
6T82
 
 | |
6T8T
 
 | |
5CZ2
 
 | | Crystal structure of a two-domain fragment of MMTV integrase | | Descriptor: | MAGNESIUM ION, Pol polyprotein, ZINC ION | | Authors: | Cook, N, Ballandras-Colas, A, Engelman, A, Cherepanov, P. | | Deposit date: | 2015-07-31 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
3GFS
 
 | | Structure of YhdA, K109D/D137K variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADPH-azoreductase | | Authors: | Staunig, N, Gruber, K. | | Deposit date: | 2009-02-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | A single intersubunit salt bridge affects oligomerization and catalytic activity in a bacterial quinone reductase
Febs J., 276, 2009
|
|
6HEL
 
 | | Structure of human USP25 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.941 Å) | | Cite: | Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.
Mol.Cell, 74, 2019
|
|
8PKA
 
 | |
2ESD
 
 | | Crystal Structure of thioacylenzyme intermediate of an Nadp Dependent Aldehyde Dehydrogenase | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase | | Authors: | D'Ambrosio, K, Didierjean, C, Benedetti, E, Aubry, A, Corbier, C. | | Deposit date: | 2005-10-26 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The first crystal structure of a thioacylenzyme intermediate in the ALDH family: new coenzyme conformation and relevance to catalysis
Biochemistry, 45, 2006
|
|
1V8O
 
 | | Crystal Structure of PAE2754 from Pyrobaculum aerophilum | | Descriptor: | CHLORIDE ION, hypothetical protein PAE2754 | | Authors: | Arcus, V.L, Backbro, K, Roos, A, Daniel, E.L, Baker, E.N. | | Deposit date: | 2004-01-12 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease
J.Biol.Chem., 279, 2004
|
|
3TCT
 
 | | Structure of wild-type TTR in complex with tafamidis | | Descriptor: | 2-(3,5-dichlorophenyl)-1,3-benzoxazole-6-carboxylic acid, Transthyretin | | Authors: | Connelly, S, Kelly, J.W, Wilson, I.A. | | Deposit date: | 2011-08-09 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3A4E
 
 | | Crystal structure of Human Transthyretin (E54G) | | Descriptor: | GLYCEROL, SULFATE ION, Transthyretin | | Authors: | Miyata, M, Sato, T, Nakamura, T, Ikemizu, S, Yamagata, Y, Kai, H. | | Deposit date: | 2009-07-06 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of the glutamic acid 54 residue in transthyretin stability and thyroxine binding
Biochemistry, 49, 2010
|
|
5XIA
 
 | | STRUCTURES OF D-XYLOSE ISOMERASE FROM ARTHROBACTER STRAIN B3728 CONTAINING THE INHIBITORS XYLITOL AND D-SORBITOL AT 2.5 ANGSTROMS AND 2.3 ANGSTROMS RESOLUTION, RESPECTIVELY | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, Xylitol | | Authors: | Henrick, K, Collyer, C.A, Blow, D.M. | | Deposit date: | 1989-07-05 | | Release date: | 1990-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of D-xylose isomerase from Arthrobacter strain B3728 containing the inhibitors xylitol and D-sorbitol at 2.5 A and 2.3 A resolution, respectively.
J.Mol.Biol., 208, 1989
|
|
