2NWP
 
 | |
4EHW
 
 | | Crystal structure of LpxK from Aquifex aeolicus at 2.3 angstrom resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Tetraacyldisaccharide 4'-kinase | | Authors: | Emptage, R.P, Daughtry, K.D, Pemble IV, C.W, Raetz, C.R.H. | | Deposit date: | 2012-04-04 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of LpxK, the 4'-kinase of lipid A biosynthesis and atypical P-loop kinase functioning at the membrane interface.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7TH1
 
 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B3 | | Descriptor: | (4S,7S,11R,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-7-benzyl-4-[(3-methylphenyl)methyl]-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
5WB6
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(11S)-11-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-6-fluoro-2-oxo-1,3,4,10,11,13-hexahydro-2H-5,9:15,12-di(azeno)-1,13-benzodiazacycloheptadecin-18-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Macrocyclic factor XIa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1YUY
 
 | | HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE GENOTYPE 2a | | Descriptor: | RNA-Dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
1YV2
 
 | | Hepatitis C virus NS5B RNA-dependent RNA Polymerase genotype 2a | | Descriptor: | GLYCEROL, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the RNA-dependent RNA Polymerase Genotype 2a of Hepatitis C Virus Reveal Two Conformations and Suggest Mechanisms of Inhibition by Non-nucleoside Inhibitors
J.Biol.Chem., 280, 2005
|
|
7V35
 
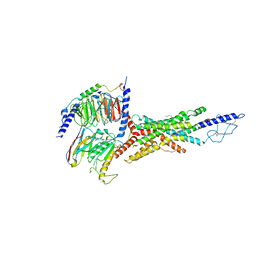 | | Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.h, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
1YVZ
 
 | | Hepatitis C Virus RNA Polymerase Genotype 2a In Complex With Non- Nucleoside Analogue Inhibitor | | Descriptor: | 3-[(2,4-DICHLOROBENZOYL)(ISOPROPYL)AMINO]-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
5FMQ
 
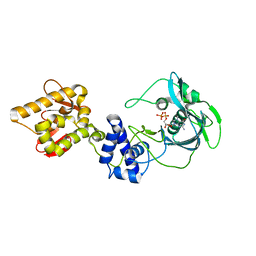 | | Crystal structure of the mid, cap-binding, mid-link and 627 domains from avian influenza A virus polymerase PB2 subunit bound to M7GTP H32 crystal form | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, INFLUENZA A PB2 SUBUNIT | | Authors: | Thierry, E, Pflug, A, Hart, D, Cusack, S. | | Deposit date: | 2015-11-07 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
7PP1
 
 | |
2NXT
 
 | |
5IPJ
 
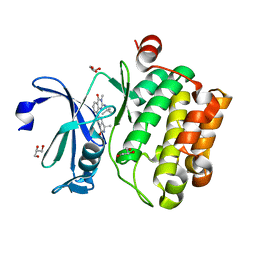 | | Crystal structure of human Pim-1 kinase in complex with a quinazolinone-pyrrolopyrrolone inhibitor. | | Descriptor: | 2-(tert-butylamino)-3-methyl-8-[(6R)-6-methyl-4-oxo-1,4,5,6-tetrahydropyrrolo[3,4-b]pyrrol-2-yl]quinazolin-4(3H)-one, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Mohr, C. | | Deposit date: | 2016-03-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Optimization of Quinazolinone-pyrrolopyrrolones as Potent and Orally Bioavailable Pan-Pim Kinase Inhibitors.
J.Med.Chem., 59, 2016
|
|
7TI2
 
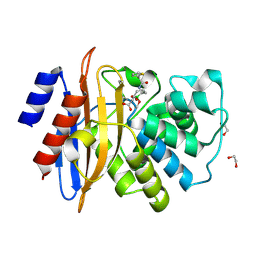 | | Structure of KPC-2 bound to RPX-7063 at 1.75A | | Descriptor: | 1,2-ETHANEDIOL, Carbapenem-hydrolyzing beta-lactamase KPC, {(3R,7S)-2-hydroxy-3-[2-(thiophen-2-yl)acetamido]-2,3,4,7-tetrahydro-1,2-oxaborepin-7-yl}acetic acid | | Authors: | Clifton, M.C, Fairman, J.W, Edwards, T.E, Hecker, S.J. | | Deposit date: | 2022-01-12 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Broad-spectrum cyclic boronate beta-lactamase inhibitors featuring an intramolecular prodrug for oral bioavailability.
Bioorg.Med.Chem., 62, 2022
|
|
5WJS
 
 | |
5YOZ
 
 | |
7SU1
 
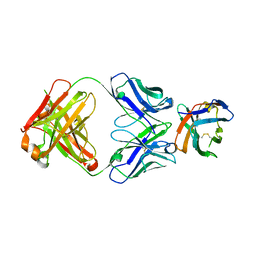 | | Crystal structure of an acidic pH-selective Ipilimumab variant Ipi.106 in complex with CTLA-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytotoxic T-lymphocyte protein 4, Fab heavy chain, ... | | Authors: | Lee, P.S, Chau, B, Strop, P. | | Deposit date: | 2021-11-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Improved therapeutic index of an acidic pH-selective antibody.
Mabs, 14, 2022
|
|
7LDE
 
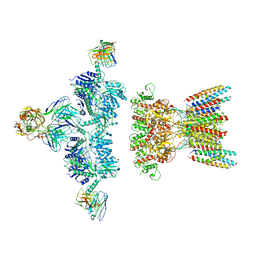 | | native AMPA receptor | | Descriptor: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | Authors: | Yu, J, Rao, P, Gouaux, E. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
7LDD
 
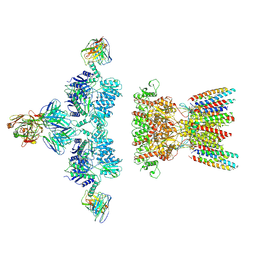 | | native AMPA receptor | | Descriptor: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | Authors: | Yu, J, Rao, P, Gouaux, E. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
7LEP
 
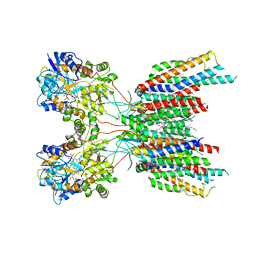 | | The composite LBD-TMD structure combined from all hippocampal AMPAR subtypes at 3.25 Angstrom resolution | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 6-[2-chloro-6-(trifluoromethoxy)phenyl]-1H-benzimidazol-2-ol, DECANE, ... | | Authors: | Yu, J, Rao, P, Gouaux, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-12 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
1XZN
 
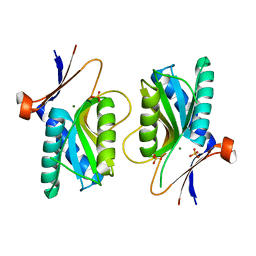 | | PYRR, THE REGULATOR OF THE PYRIMIDINE BIOSYNTHETIC OPERON IN BACILLUS CALDOLYTICUS, sulfate-bound form | | Descriptor: | MAGNESIUM ION, PyrR bifunctional protein, SULFATE ION | | Authors: | Chander, P, Halbig, K.M, Miller, J.K, Fields, C.J, Bonner, H.K, Grabner, G.K, Switzer, R.L, Smith, J.L. | | Deposit date: | 2004-11-12 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of the Nucleotide Complex of PyrR, the pyr Attenuation Protein from Bacillus caldolyticus, Suggests Dual Regulation by Pyrimidine and Purine Nucleotides.
J.Bacteriol., 187, 2005
|
|
7SU0
 
 | | Crystal structure of an acidic pH-selective Ipilimumab variant Ipi.105 in complex with CTLA-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, Cytotoxic T-lymphocyte protein 4, ... | | Authors: | Lee, P.S, Chau, B, Strop, P. | | Deposit date: | 2021-11-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Improved therapeutic index of an acidic pH-selective antibody.
Mabs, 14, 2022
|
|
6E0T
 
 | | C-terminal domain of Fission Yeast OFD1 | | Descriptor: | Prolyl 3,4-dihydroxylase ofd1 | | Authors: | Bianchet, M.A, Amzel, L.M, Espenshade, P.J, Yeh, T. | | Deposit date: | 2018-07-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The hypoxic regulator of sterol synthesis nro1 is a nuclear import adaptor.
Structure, 19, 2011
|
|
4BIM
 
 | |
1XZ8
 
 | | Pyrr, The Regulator Of The Pyrimidine Biosynthetic Operon In Bacillus caldolyticus, Nucleotide-bound form | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chander, P, Halbig, K.M, Miller, J.K, Fields, C.J, Bonner, H.K, Grabner, G.K, Switzer, R.L, Smith, J.L. | | Deposit date: | 2004-11-11 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Nucleotide Complex of PyrR, the pyr Attenuation Protein from Bacillus caldolyticus, Suggests Dual Regulation by Pyrimidine and Purine Nucleotides.
J.Bacteriol., 187, 2005
|
|
5Z7Z
 
 | | Crystal structure of Striga hermonthica Dwarf14 (ShD14) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dwarf 14, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
