6LFU
 
 | | Poa1p F152A mutant in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.123 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LFT
 
 | | Poa1p S30A mutant in complex with ADP-ribose | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LFS
 
 | | Poa1p H23A mutant in complex with ADP-ribose | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LFQ
 
 | | Crystal structure of Poa1p in apo form | | Descriptor: | ADP-ribose 1''-phosphate phosphatase, GLYCEROL | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LFR
 
 | | Poa1p in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LH4
 
 | | Crystal structural of MacroD1-ADPr complex | | Descriptor: | ADP-ribose glycohydrolase MACROD1, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Yang, X, Ma, Y, Li, Y. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Molecular basis for the MacroD1-mediated hydrolysis of ADP-ribosylation.
DNA Repair (Amst), 94, 2020
|
|
6MEB
 
 | | Crystal structure of Tylonycteris bat coronavirus HKU4 macrodomain in complex with nicotinamide adenine dinucleotide (NAD+) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
6MEA
 
 | | Crystal structure of a Tylonycteris bat coronavirus HKU4 macrodomain in complex with adenosine diphosphate ribose (ADP-ribose) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
6MEN
 
 | | Crystal structure of a Tylonycteris bat coronavirus HKU4 macrodomain in complex with adenosine diphosphate glucose (ADP-glucose) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
5KIV
 
 | | Crystal structure of SauMacro (SAV0325) | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Protein-ADP-ribose hydrolase, ... | | Authors: | Williams, R.S, Appel, C.D, Feld, G.K, Wallace, B.D. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the sirtuin-linked macrodomain SAV0325 from Staphylococcus aureus.
Protein Sci., 25, 2016
|
|
5LAU
 
 | | Oceanobacillus iheyensis macrodomain mutant G37V with ADPR | | Descriptor: | GLYCEROL, MacroD-type macrodomain, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
5LBP
 
 | | Oceanobacillus iheyensis macrodomain mutant N30A | | Descriptor: | MacroD-type macrodomain, PHOSPHATE ION | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-16 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
5L9Q
 
 | | OCEANOBACILLUS IHEYENSIS MACRODOMAIN WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MACROD-TYPE MACRODOMAIN, SULFATE ION | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-10 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
5LNC
 
 | |
5LW0
 
 | | Oryza sativa APL macrodomain in complex with ADP-ribose | | Descriptor: | Basic helix-loop-helix, putative, expressed, ... | | Authors: | Ariza, A. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The role of ADP-ribosylation in regulating DNA interstrand crosslink repair.
J.Cell.Sci., 129, 2016
|
|
5M3I
 
 | | Macrodomain of Mycobacterium tuberculosis DarG | | Descriptor: | CHLORIDE ION, RNase III inhibitor | | Authors: | Ariza, A. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA.
Mol. Cell, 64, 2016
|
|
5L9K
 
 | | OCEANOBACILLUS IHEYENSIS MACRODOMAIN WITH ADPR | | Descriptor: | GLYCEROL, MACROD-TYPE MACRODOMAIN, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-10 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
5LCC
 
 | | Oceanobacillus iheyensis macrodomain mutant D40A | | Descriptor: | MACROD-TYPE MACRODOMAIN | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
5M3E
 
 | | Macrodomain of Thermus aquaticus DarG in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Appr-1-p processing domain protein, CHLORIDE ION | | Authors: | Ariza, A. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA.
Mol. Cell, 64, 2016
|
|
5M31
 
 | | Macrodomain of Thermus aquaticus DarG | | Descriptor: | Appr-1-p processing domain protein, CHLORIDE ION, GLYCEROL | | Authors: | Ariza, A. | | Deposit date: | 2016-10-13 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA.
Mol. Cell, 64, 2016
|
|
6W8K
 
 | | Co-crystal structures of CHIKV nsP3 macrodomain with pyrimidone fragments | | Descriptor: | 5,6-dimethyl-2-oxo-2,3-dihydropyrimidine-4-carboxylic acid, Nonstructural polyprotein | | Authors: | Zhang, S, Garzan, A, Augelli-Szafran, C.E, Pathak, A.K, Wu, M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pyrimidone inhibitors targeting Chikungunya Virus nsP3 macrodomain by fragment-based drug design.
Plos One, 16, 2021
|
|
6W8M
 
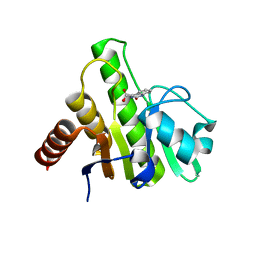 | | Co-crystal structures of CHIKV nsP3 macrodomain with pyrimidone fragments | | Descriptor: | 2-oxo-1,2-dihydroquinazoline-4-carboxylic acid, Nonstructural polyprotein | | Authors: | Zhang, S, Garzan, A, Pathak, A.K, Augelli-Szafran, C.E, Wu, M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Pyrimidone inhibitors targeting Chikungunya Virus nsP3 macrodomain by fragment-based drug design.
Plos One, 16, 2021
|
|
6W8Q
 
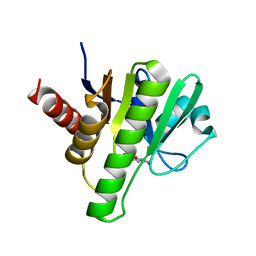 | | Co-crystal structures of CHIKV nsP3 macrodomain with pyrimidone fragments | | Descriptor: | 2-oxo-1,2,5,6,7,8-hexahydroquinazoline-4-carboxylic acid, Nonstructural polyprotein | | Authors: | Zhang, S, Garzan, A, Pathak, A.K, Augelli-Szafran, C.E, Wu, M. | | Deposit date: | 2020-03-21 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Pyrimidone inhibitors targeting Chikungunya Virus nsP3 macrodomain by fragment-based drug design.
Plos One, 16, 2021
|
|
6VUQ
 
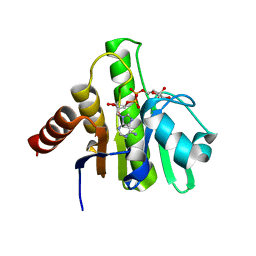 | | Crystal structure of CHIKV nsP3 macrodomain soaked with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Nonstructural polyprotein | | Authors: | Wu, M. | | Deposit date: | 2020-02-16 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Pyrimidone inhibitors targeting Chikungunya Virus nsP3 macrodomain by fragment-based drug design.
Plos One, 16, 2021
|
|
6W02
 
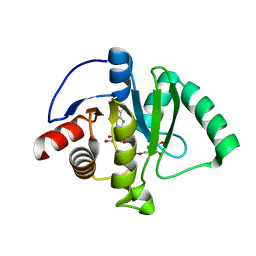 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in the complex with ADP ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-28 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
