5WAG
 
 | | ADC-7 in complex with boronic acid transition state inhibitor S06017 | | Descriptor: | 1-{[hydroxy(phosphonooxy)boranyl]methyl}-1H-1,2,3-triazole-4-carboxylic acid, Beta-lactamase, SUCCINIC ACID | | Authors: | Powers, R.A, Wallar, B.J. | | Deposit date: | 2017-06-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.931 Å) | | Cite: | Structure-Based Analysis of Boronic Acids as Inhibitors of Acinetobacter-Derived Cephalosporinase-7, a Unique Class C beta-Lactamase.
ACS Infect Dis, 4, 2018
|
|
8AIK
 
 | |
8AKH
 
 | | Crystal structure of DltE from L. plantarum soaked with LTA | | Descriptor: | Beta-lactamase family protein, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Ravaud, S, Nikolopoulos, N, Grangeasse, C. | | Deposit date: | 2022-07-29 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function analysis of Lactiplantibacillus plantarum DltE& reveals D-alanylated lipoteichoic acids as direct cues supporting Drosophila juvenile growth.
Elife, 12, 2023
|
|
8AGR
 
 | | Crystal structure of DltE from L. plantarum, apo form | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase family protein, SULFATE ION | | Authors: | Ravaud, S, Nikolopoulos, N, Grangeasse, C. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-function analysis of Lactiplantibacillus plantarum DltE& reveals D-alanylated lipoteichoic acids as direct cues supporting Drosophila juvenile growth.
Elife, 12, 2023
|
|
8AJI
 
 | | Crystal structure of DltE from L. plantarum, TCEP form | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Beta-lactamase family protein, GLYCEROL, ... | | Authors: | Ravaud, S, Nikolopoulos, N, Grangeasse, C. | | Deposit date: | 2022-07-28 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-function analysis of Lactiplantibacillus plantarum DltE& reveals D-alanylated lipoteichoic acids as direct cues supporting Drosophila juvenile growth.
Elife, 12, 2023
|
|
5WAE
 
 | | ADC-7 in complex with boronic acid transition state inhibitor CR167 | | Descriptor: | 3-(5,7,7-trihydroxy-2,2,7-trioxo-6-oxa-2lambda~6~-thia-3-aza-7lambda~5~-phospha-5-boraheptan-1-yl)benzoic acid, Beta-lactamase | | Authors: | Powers, R.A, Wallar, B.J. | | Deposit date: | 2017-06-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structure-Based Analysis of Boronic Acids as Inhibitors of Acinetobacter-Derived Cephalosporinase-7, a Unique Class C beta-Lactamase.
ACS Infect Dis, 4, 2018
|
|
5WAF
 
 | | ADC-7 in complex with boronic acid transition state inhibitor CR192 | | Descriptor: | Beta-lactamase, phosphonooxy-[[[4-(1~{H}-1,2,3,4-tetrazol-5-yl)-2-(trifluoromethyl)phenyl]sulfonylamino]methyl]borinic acid | | Authors: | Powers, R.A, Wallar, B.J. | | Deposit date: | 2017-06-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Based Analysis of Boronic Acids as Inhibitors of Acinetobacter-Derived Cephalosporinase-7, a Unique Class C beta-Lactamase.
ACS Infect Dis, 4, 2018
|
|
5W12
 
 | |
5W13
 
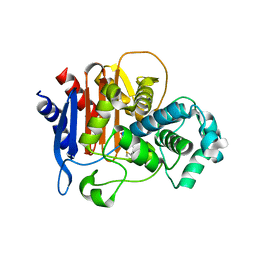 | |
5WAD
 
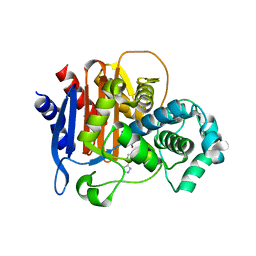 | | ADC-7 in complex with boronic acid transition state inhibitor CR161 | | Descriptor: | Beta-lactamase, PHOSPHATE ION, phosphonooxy-[[[6-(1~{H}-1,2,3,4-tetrazol-5-yl)pyridin-3-yl]sulfonylamino]methyl]borinic acid | | Authors: | Powers, R.A, Wallar, B.J. | | Deposit date: | 2017-06-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-Based Analysis of Boronic Acids as Inhibitors of Acinetobacter-Derived Cephalosporinase-7, a Unique Class C beta-Lactamase.
ACS Infect Dis, 4, 2018
|
|
5ZFL
 
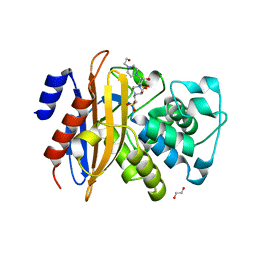 | | Crystal structure of beta-lactamase PenP mutant E166Y | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase | | Authors: | Pan, X, Zhao, Y. | | Deposit date: | 2018-03-06 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The hydrolytic water molecule of Class A beta-lactamase relies on the acyl-enzyme intermediate ES* for proper coordination and catalysis.
Sci Rep, 10, 2020
|
|
5ZG6
 
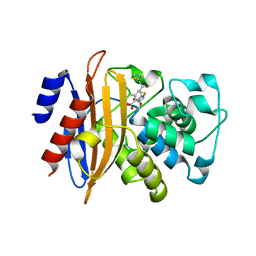 | |
5ZH8
 
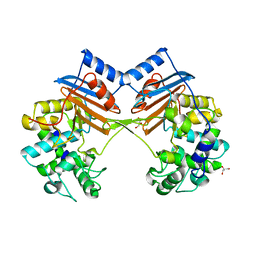 | | Crystal Structure of FmtA from Staphylococcus aureus at 2.58 A | | Descriptor: | GLYCEROL, Protein FmtA | | Authors: | Dalal, V, Kumar, P, Golemi-Kotra, D, Kumar, P. | | Deposit date: | 2018-03-12 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Repurposing an Ancient Protein Core Structure: Structural Studies on FmtA, a Novel Esterase of Staphylococcus aureus.
J.Mol.Biol., 431, 2019
|
|
1BLS
 
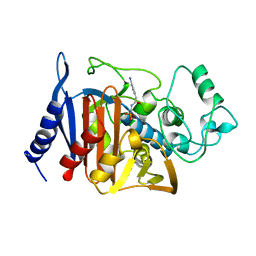 | |
2DCF
 
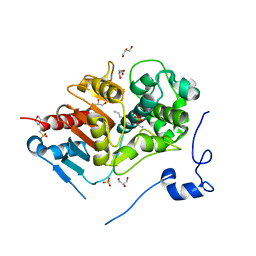 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/H266N mutant with substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2006-01-06 | | Release date: | 2007-01-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Nylon-oligomer degrading enzyme/substrate complex: catalytic mechanism of 6-aminohexanoate-dimer hydrolase
J.Mol.Biol., 370, 2007
|
|
2DRW
 
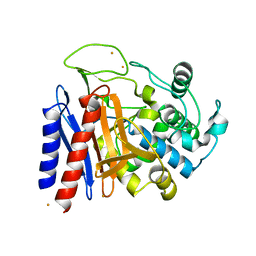 | | The crystal structutre of D-amino acid amidase from Ochrobactrum anthropi SV3 | | Descriptor: | BARIUM ION, D-Amino acid amidase | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Functional Characterization of a D-Stereospecific Amino Acid Amidase from Ochrobactrum anthropi SV3, a New Member of the Penicillin-recognizing Proteins
J.Mol.Biol., 368, 2007
|
|
2DNS
 
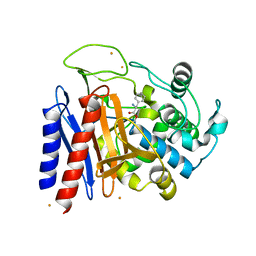 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with D-Phenylalanine | | Descriptor: | BARIUM ION, D-PHENYLALANINE, D-amino acid amidase | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Functional Characterization of a D-Stereospecific Amino Acid Amidase from Ochrobactrum anthropi SV3, a New Member of the Penicillin-recognizing Proteins
J.Mol.Biol., 368, 2007
|
|
2E8I
 
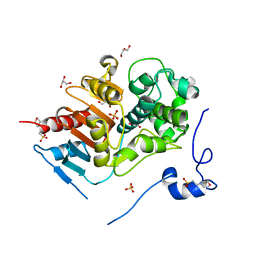 | | Structure of 6-aminohexanoate-dimer hydrolase, D1 mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Shibata, N, Higuchi, Y, Negoro, S. | | Deposit date: | 2007-01-20 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold.
Febs J., 276, 2009
|
|
8J5V
 
 | |
2EFX
 
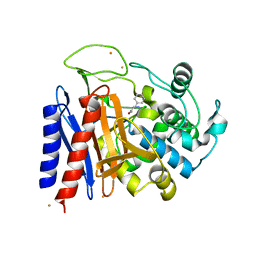 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with L-phenylalanine amide | | Descriptor: | BARIUM ION, D-amino acid amidase, PHENYLALANINE AMIDE | | Authors: | Okazaki, S, Suzuki, A, Mizushima, T, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of D-amino-acid amidase complexed with L-phenylalanine and with L-phenylalanine amide: insight into the D-stereospecificity of D-amino-acid amidase from Ochrobactrum anthropi SV3.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2EFU
 
 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with L-phenylalanine | | Descriptor: | BARIUM ION, D-Amino acid amidase, PHENYLALANINE | | Authors: | Okazaki, S, Suzuki, A, Mizushima, T, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of D-amino-acid amidase complexed with L-phenylalanine and with L-phenylalanine amide: insight into the D-stereospecificity of D-amino-acid amidase from Ochrobactrum anthropi SV3.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
8KEM
 
 | | PKS domains-fused AmpC EC2 | | Descriptor: | SDR family NAD(P)-dependent oxidoreductase,Beta-lactamase | | Authors: | Son, S.Y, Bae, D.W, Cha, S.S. | | Deposit date: | 2023-08-12 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.76998281 Å) | | Cite: | Structural investigation of the docking domain assembly from trans-AT polyketide synthases.
Structure, 32, 2024
|
|
8JB7
 
 | | Crystal structure of CMY-185 | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2023-05-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into the molecular mechanism of high-level ceftazidime-avibactam resistance conferred by CMY-185.
Mbio, 15, 2024
|
|
8JB8
 
 | | Crystal structure of CMY-185 complex with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2023-05-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the molecular mechanism of high-level ceftazidime-avibactam resistance conferred by CMY-185.
Mbio, 15, 2024
|
|
2BLM
 
 | |
