3HAS
 
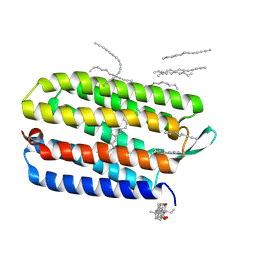 | | Crystal structure of bacteriorhodopsin mutant L152A crystallized from bicelles | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Bacteriorhodopsin, DECANE, ... | | Authors: | Joh, N.H, Yang, D, Bowie, J.U. | | Deposit date: | 2009-05-02 | | Release date: | 2009-09-22 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Similar energetic contributions of packing in the core of membrane and water-soluble proteins.
J.Am.Chem.Soc., 131, 2009
|
|
2QX8
 
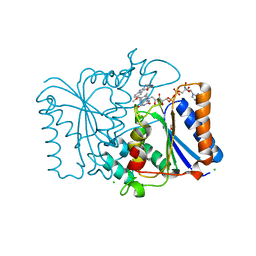 | | Crystal Structure of Quinone Reductase II | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Calamini, B, Santarsiero, B.D, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2007-08-10 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Biochem.J., 413, 2008
|
|
3H9Q
 
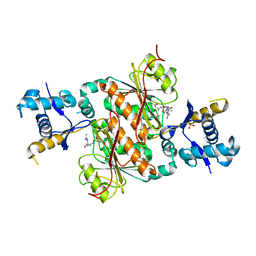 | | Crystal structure of E. coli MccB + SeMet MccA | | Descriptor: | MccB protein, Microcin C7 ANALOG, SULFATE ION, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3U31
 
 | |
2QZB
 
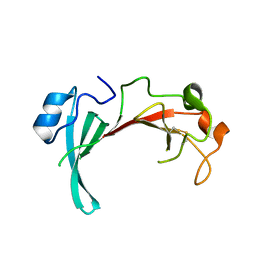 | | Crystal structure of the uncharacterized protein yfeY from Escherichia coli | | Descriptor: | Uncharacterized protein yfeY | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Wu, B, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-16 | | Release date: | 2007-08-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the uncharacterized protein yfeY from Escherichia coli.
To be Published
|
|
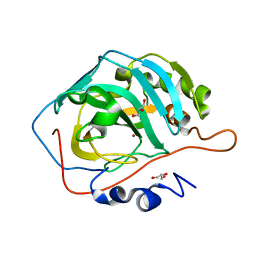
3U3A
 
 | |
3U3J
 
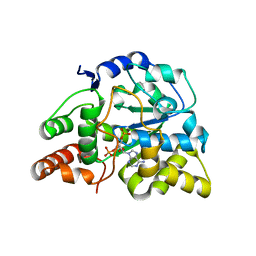 | | Crystal structure of hSULT1A1 bound to PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase 1A1 | | Authors: | Guttman, C, Berger, I, Aharoni, A, Zarivach, R. | | Deposit date: | 2011-10-06 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The molecular basis for the broad substrate specificity of human sulfotransferase 1A1.
Plos One, 6, 2011
|
|
3U41
 
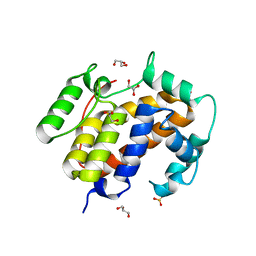 | |
3HD2
 
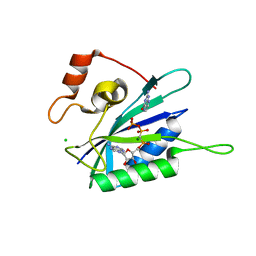 | | Crystal structure of E. coli HPPK(Q50A) in complex with MgAMPCPP and pterin | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-05-06 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Role of loop coupling in enzymatic catalysis and conformational dynamics
To be Published
|
|
3HE1
 
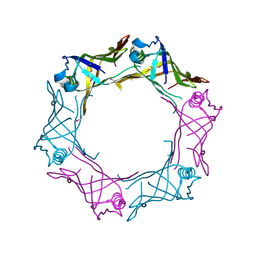 | | Secreted protein Hcp3 from Pseudomonas aeruginosa. | | Descriptor: | GLYCEROL, Major exported Hcp3 protein | | Authors: | Osipiuk, J, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-07 | | Release date: | 2009-06-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Crystal structure of secretory protein Hcp3 from Pseudomonas aeruginosa.
J.Struct.Funct.Genom., 12, 2011
|
|
3HEQ
 
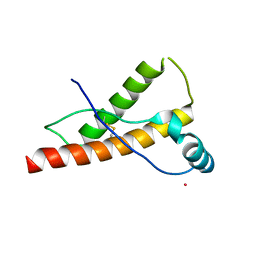 | | Human prion protein variant D178N with M129 | | Descriptor: | CADMIUM ION, Major prion protein | | Authors: | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | Deposit date: | 2009-05-10 | | Release date: | 2010-01-12 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3TZA
 
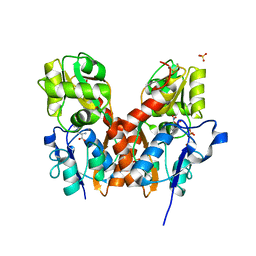 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (S)-2-amino-3-(2-(2-carboxyethyl)-5-chloro-4-nitrophenyl)propionic acid at 1.9A resolution | | Descriptor: | (S)-2-amino-3-(2-(2-carboxyethyl)-5-chloro-4-nitrophenyl)propionic acid, Glutamate receptor 2,Glutamate receptor 2, SULFATE ION | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2011-09-27 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new phenylalanine derivative acts as an antagonist at the AMPA receptor GluA2 and introduces partial domain closure: synthesis, resolution, pharmacology, and crystal structure
J.Med.Chem., 54, 2011
|
|
3HG4
 
 | | Human alpha-galactosidase catalytic mechanism 3. Covalent intermediate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guce, A.I, Clark, N.E, Garman, S.C. | | Deposit date: | 2009-05-13 | | Release date: | 2009-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic mechanism of human alpha-galactosidase.
J.Biol.Chem., 285, 2010
|
|
3HGG
 
 | | Crystal Structure of CmeR Bound to Cholic Acid | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Routh, M.D, Yang, F. | | Deposit date: | 2009-05-13 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for anionic ligand recognition by multidrug
binding proteins: Crystal structures of CmeR-bile acid complexes
To be Published
|
|
3HGW
 
 | | Apo Structure of Pseudomonas aeruginosa Isochorismate-Pyruvate Lyase I87T mutant | | Descriptor: | CALCIUM ION, Salicylate biosynthesis protein pchB | | Authors: | Luo, Q, Lamb, A.L. | | Deposit date: | 2009-05-14 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-function analyses of isochorismate-pyruvate lyase from Pseudomonas aeruginosa suggest differing catalytic mechanisms for the two pericyclic reactions of this bifunctional enzyme
Biochemistry, 48, 2009
|
|
3HIQ
 
 | |
3TQY
 
 | | Structure of a single-stranded DNA-binding protein (ssb), from Coxiella burnetii | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.6001 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
2R5P
 
 | | Crystal Structure Analysis of HIV-1 Subtype C Protease Complexed with Indinavir | | Descriptor: | CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, Protease, ... | | Authors: | Coman, R.M, Robbins, A.H, McKenna, R, Dunn, B.M. | | Deposit date: | 2007-09-04 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Contribution of Naturally Occurring Polymorphisms in Altering the Biochemical and Structural Characteristics of HIV-1 Subtype C Protease
Biochemistry, 47, 2008
|
|
3TRF
 
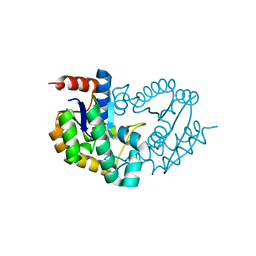 | | Structure of a shikimate kinase (aroK) from Coxiella burnetii | | Descriptor: | SULFATE ION, Shikimate kinase | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3U3M
 
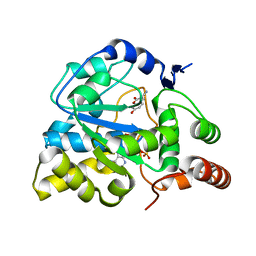 | | Crystal structure of Human SULT1A1 bound to PAP and 3-Cyano-7-hydroxycoumarin | | Descriptor: | 7-hydroxy-2-oxo-2H-chromene-3-carbonitrile, ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase 1A1 | | Authors: | Guttman, C, Berger, I, Aharoni, A, Zarivach, R. | | Deposit date: | 2011-10-06 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular basis for the broad substrate specificity of human sulfotransferase 1A1.
Plos One, 6, 2011
|
|
3TRV
 
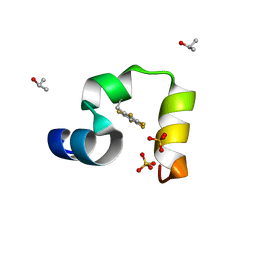 | | Crystal structure of quasiracemic villin headpiece subdomain containing (F5Phe17) substitution | | Descriptor: | D-Villin-1, ISOPROPYL ALCOHOL, L-Villin-1, ... | | Authors: | Mortenson, D.E, Satyshur, K.A, Gellman, S.H, Forest, K.T. | | Deposit date: | 2011-09-11 | | Release date: | 2012-01-25 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Quasiracemic crystallization as a tool to assess the accommodation of noncanonical residues in nativelike protein conformations.
J.Am.Chem.Soc., 134, 2012
|
|
3TRY
 
 | | Crystal structure of racemic villin headpiece subdomain in space group I-4c2 | | Descriptor: | D-Villin-1, SULFATE ION | | Authors: | Mortenson, D.E, Satyshur, K.A, Gellman, S.H, Forest, K.T. | | Deposit date: | 2011-09-11 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quasiracemic crystallization as a tool to assess the accommodation of noncanonical residues in nativelike protein conformations.
J.Am.Chem.Soc., 134, 2012
|
|
3HG5
 
 | | Human alpha-galactosidase catalytic mechanism 4. Product bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, Alpha-galactosidase A, ... | | Authors: | Guce, A.I, Clark, N.E, Garman, S.C. | | Deposit date: | 2009-05-13 | | Release date: | 2009-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic mechanism of human alpha-galactosidase.
J.Biol.Chem., 285, 2010
|
|
3HIV
 
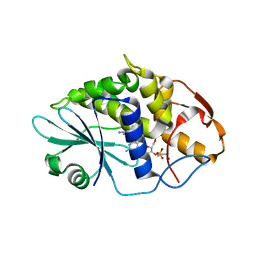 | | Crystal structure of Saporin-L1 in complex with the trinucleotide inhibitor, a transition state analogue | | Descriptor: | (2R,3R,4R,5R)-5-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)-2-({[(S)-({(3R,4R)-4-({[(S)-{[(2R,3R,4R,5R)-5-(2-amino-6-oxo-6,8-dihydro-9H-purin-9-yl)-2-(hydroxymethyl)-4-methoxytetrahydrofuran-3-yl]oxy}(hydroxy)phosphoryl]oxy}methyl)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]pyrrolidin-3-yl}oxy)(hydroxy)phosphoryl]oxy}methyl)-4-methoxytetrahydrofuran-3-yl 3-hydroxypropyl hydrogen (S)-phosphate, Vacuolar saporin | | Authors: | Ho, M, Sturm, M.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HJ6
 
 | | Structure of Halothermothrix orenii fructokinase (FRK) | | Descriptor: | Fructokinase | | Authors: | Chua, T.K, Seetharaman, J, Kasprzak, J.M, Ng, C, Patel, B.K, Love, C, Bujnicki, J.M, Sivaraman, J. | | Deposit date: | 2009-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a fructokinase homolog from Halothermothrix orenii
J.Struct.Biol., 171, 2010
|
|
