1TIN
 
 | |
1TAN
 
 | | TANDEM DNA, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(P*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*CP*A)-3'), DNA (5'-D(P*TP*GP*GP*AP*GP*CP*TP*G)-3') | | Authors: | Denisov, A, Sandstrom, A, Maltseva, T, Pyshnyi, D, Ivanova, E, Zarytova, V, Chattopadhyaya, J. | | Deposit date: | 1997-06-17 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of estrone (Es)-tethered tandem DNA duplex: [d(5'pCAGCp3')-Es] + [Es-d(5'pTCCA3')]: d(5'pTGGAGCTG3').
J.Biomol.Struct.Dyn., 15, 1997
|
|
1TFI
 
 | | A NOVEL ZN FINGER MOTIF IN THE BASAL TRANSCRIPTIONAL MACHINERY: THREE-DIMENSIONAL NMR STUDIES OF THE NUCLEIC-ACID BINDING DOMAIN OF TRANSCRIPTIONAL ELONGATION FACTOR TFIIS | | Descriptor: | TRANSCRIPTIONAL ELONGATION FACTOR SII, ZINC ION | | Authors: | Qian, X, Gozani, S, Yoon, H.S, Jeon, C.J, Agarwal, K, Weiss, M.A. | | Deposit date: | 1993-04-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Novel zinc finger motif in the basal transcriptional machinery: three-dimensional NMR studies of the nucleic acid binding domain of transcriptional elongation factor TFIIS.
Biochemistry, 32, 1993
|
|
1TCG
 
 | |
1WHF
 
 | | COAGULATION FACTOR, NMR, 15 STRUCTURES | | Descriptor: | COAGULATION FACTOR X | | Authors: | Sunnerhagen, M, Olah, G.A, Stenflo, J, Forsen, S, Drakenberg, T, Trewhella, J. | | Deposit date: | 1996-06-18 | | Release date: | 1997-05-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | The relative orientation of Gla and EGF domains in coagulation factor X is altered by Ca2+ binding to the first EGF domain. A combined NMR-small angle X-ray scattering study.
Biochemistry, 35, 1996
|
|
1WHE
 
 | | COAGULATION FACTOR, NMR, 20 STRUCTURES | | Descriptor: | COAGULATION FACTOR X | | Authors: | Sunnerhagen, M, Olah, G.A, Stenflo, J, Forsen, S, Drakenberg, T, Trewhella, J. | | Deposit date: | 1996-06-18 | | Release date: | 1997-05-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | The relative orientation of Gla and EGF domains in coagulation factor X is altered by Ca2+ binding to the first EGF domain. A combined NMR-small angle X-ray scattering study.
Biochemistry, 35, 1996
|
|
2KNR
 
 | | Solution structure of protein Atu0922 from A. tumefaciens. Northeast Structural Genomics Consortium target AtT13. Ontario Center for Structural Proteomics target ATC0905 | | Descriptor: | Uncharacterized protein ATC0905 | | Authors: | Gutmanas, A, Yee, S, Lemak, A, Fares, A, Semesi, A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein atc0905 from A. tumefaciens
To be Published
|
|
2JM4
 
 | |
1QDF
 
 | | THE NMR STUDY OF DNA QUADRUPLEX STRUCTURE, APTAMER (15MER) DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Marathias, V.M, Wang, K.Y, Kumar, S, Swaminathan, S, Bolton, P.H. | | Deposit date: | 1996-04-11 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the number and location of the manganese binding sites of DNA quadruplexes in solution by EPR and NMR in the presence and absence of thrombin.
J.Mol.Biol., 260, 1996
|
|
1QDI
 
 | | THE NMR STUDY OF DNA QUADRUPLEX STRUCTURE, (12MER) DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3') | | Authors: | Marathias, V.M, Wang, K.Y, Kumar, S, Swaminathan, S, Bolton, P.H. | | Deposit date: | 1996-04-11 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the number and location of the manganese binding sites of DNA quadruplexes in solution by EPR and NMR in the presence and absence of thrombin.
J.Mol.Biol., 260, 1996
|
|
1QDH
 
 | | THE NMR STUDY OF DNA QUADRUPLEX STRUCTURE, APTAMER (15MER) DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), MANGANESE (II) ION | | Authors: | Marathias, V.M, Wang, K.Y, Kumar, S, Swaminathan, S, Bolton, P.H. | | Deposit date: | 1996-04-11 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the number and location of the manganese binding sites of DNA quadruplexes in solution by EPR and NMR in the presence and absence of thrombin.
J.Mol.Biol., 260, 1996
|
|
1QDK
 
 | | THE NMR STUDY OF DNA QUADRUPLEX STRUCTURE, (12MER) DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3'), MANGANESE (II) ION | | Authors: | Marathias, V.M, Wang, K.Y, Kumar, S, Swaminathan, S, Bolton, P.H. | | Deposit date: | 1996-04-11 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the number and location of the manganese binding sites of DNA quadruplexes in solution by EPR and NMR in the presence and absence of thrombin.
J.Mol.Biol., 260, 1996
|
|
1QET
 
 | | TANDEM GU MISMATCHES IN RNA, NMR, 30 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*AP*UP*GP*UP*CP*C)-3') | | Authors: | Mcdowell, J.A, He, L, Chen, X, Turner, D.H. | | Deposit date: | 1997-03-04 | | Release date: | 1997-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Investigation of the structural basis for thermodynamic stabilities of tandem GU wobble pairs: NMR structures of (rGGAGUUCC)2 and (rGGAUGUCC)2.
Biochemistry, 36, 1997
|
|
1R9P
 
 | | Solution NMR Structure Of The Haemophilus Influenzae Iron-Sulfur Cluster Assembly Protein U (IscU) with Zinc Bound at the Active Site. Northeast Structural Genomics Consortium Target IR24. | | Descriptor: | NifU-like protein, ZINC ION | | Authors: | Ramelot, T.A, Cort, J.R, Xiao, R, Shastry, R, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-10-30 | | Release date: | 2004-11-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site.
J.Mol.Biol., 344, 2004
|
|
1RTN
 
 | |
1RHT
 
 | | 24-MER RNA HAIRPIN COAT PROTEIN BINDING SITE FOR BACTERIOPHAGE R17 (NMR, MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | RNA (5'-R(P*GP*GP*GP*AP*CP*UP*GP*AP*CP*GP*AP*UP*CP*AP*CP*GP*CP*AP*GP*UP*CP*UP*AP*U)-3') | | Authors: | Borer, P.N, Lin, Y, Wang, S, Roggenbuck, M.W, Gott, J.M, Uhlenbeck, O.C, Pelczer, I. | | Deposit date: | 1995-03-03 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Proton NMR and structural features of a 24-nucleotide RNA hairpin.
Biochemistry, 34, 1995
|
|
1TCJ
 
 | |
1TCK
 
 | |
2D8B
 
 | | Solution structure of the second tandem cofilin-domain of mouse twinfilin | | Descriptor: | Twinfilin-1 | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Sato, M, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-02 | | Release date: | 2006-06-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains
Protein Sci., 18, 2009
|
|
2HZ8
 
 | | QM/MM structure refined from NMR-structure of a single chain diiron protein | | Descriptor: | De novo designed diiron protein, ZINC ION | | Authors: | Calhoun, J.R, Liu, W, Spiegel, K, Dal Peraro, M, Klein, M.L, Wand, A.J, DeGrado, W.F. | | Deposit date: | 2006-08-08 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a designed metalloprotein and complementary molecular dynamics refinement.
Structure, 16, 2008
|
|
2JSV
 
 | | Dipole tensor-based refinement for atomic-resolution structure determination of a nanocrystalline protein by solid-state NMR spectroscopy | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Franks, W, Wylie, B.J, Frericks, H.L, Nieuwkoop, A.J, Mayrhofer, R, Shah, G.J, Graesser, D.T, Rienstra, C.M. | | Deposit date: | 2007-07-16 | | Release date: | 2008-04-15 | | Last modified: | 2024-05-29 | | Method: | SOLID-STATE NMR | | Cite: | Dipole tensor-based atomic-resolution structure determination of a nanocrystalline protein by solid-state NMR
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1QJL
 
 | | METALLOTHIONEIN MTA FROM SEA URCHIN (BETA DOMAIN) | | Descriptor: | CADMIUM ION, METALLOTHIONEIN | | Authors: | Riek, R, Precheur, B, Wang, Y, Mackay, E.A, Wider, G, Guntert, P, Liu, A, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the sea urchin (Strongylocentrotus purpuratus) metallothionein MTA.
J. Mol. Biol., 291, 1999
|
|
1YN1
 
 | |
1STU
 
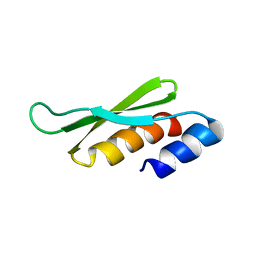 | | DOUBLE STRANDED RNA BINDING DOMAIN | | Descriptor: | MATERNAL EFFECT PROTEIN STAUFEN | | Authors: | Bycroft, M. | | Deposit date: | 1995-05-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5.
EMBO J., 14, 1995
|
|
1YN2
 
 | |
