1IY3
 
 | | Solution Structure of the Human lysozyme at 4 degree C | | Descriptor: | Lysozyme | | Authors: | Kumeta, H, Miura, A, Kobashigawa, Y, Miura, K, Oka, C, Nitta, K, Nemoto, N, Tsuda, S. | | Deposit date: | 2002-07-15 | | Release date: | 2002-07-31 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in human lysozyme elucidated by three-dimensional NMR spectroscopy
Biochemistry, 42, 2003
|
|
1IOT
 
 | | STABILIZATION OF HEN EGG WHITE LYSOZYME BY A CAVITY-FILLING MUTATION | | Descriptor: | LYSOZYME C | | Authors: | Ohmura, T, Ueda, T, Ootsuka, K, Saito, M, Imoto, T. | | Deposit date: | 2001-03-28 | | Release date: | 2001-04-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Stabilization of hen egg white lysozyme by a cavity-filling mutation.
Protein Sci., 10, 2001
|
|
1IEE
 
 | | STRUCTURE OF TETRAGONAL HEN EGG WHITE LYSOZYME AT 0.94 A FROM CRYSTALS GROWN BY THE COUNTER-DIFFUSION METHOD | | Descriptor: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | Authors: | Sauter, C, Otalora, F, Gavira, J.-A, Vidal, O, Giege, R, Garcia-Ruiz, J.-M. | | Deposit date: | 2001-04-09 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Structure of tetragonal hen egg-white lysozyme at 0.94 A from crystals grown by the counter-diffusion method.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IOC
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME, EAEA-I56T | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Goda, S, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-02-27 | | Release date: | 2002-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Elongation in a beta-structure promotes amyloid-like fibril formation of human lysozyme.
J.Biochem., 132, 2002
|
|
1IR8
 
 | | IM mutant of lysozyme | | Descriptor: | lysozyme | | Authors: | Ohmura, T, Ueda, T, Hashimoto, Y, Imoto, T. | | Deposit date: | 2001-09-19 | | Release date: | 2001-10-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Tolerance of point substitution of methionine for isoleucine in hen egg white lysozyme.
Protein Eng., 14, 2001
|
|
1SQ2
 
 | | Crystal Structure Analysis of the Nurse Shark New Antigen Receptor (NAR) Variable Domain in Complex With Lysozyme | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Stanfield, R.L, Dooley, H, Flajnik, M.F, Wilson, I.A. | | Deposit date: | 2004-03-17 | | Release date: | 2004-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a shark single-domain antibody V region in complex with lysozyme.
Science, 305, 2004
|
|
6P4B
 
 | | HyHEL10 fab variant HyHEL10-4x (heavy chain mutations L4F, Y33H, S56N, and Y58F) bound to hen egg lysozyme variant HEL2x-flex (mutations R21Q, R73E, C76S, and C94S) | | Descriptor: | CHLORIDE ION, HyHEL10 Fab heavy chain, HyHEL10 Fab light chain, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4A
 
 | |
3F6Z
 
 | |
3G3A
 
 | |
3G3B
 
 | |
7Q06
 
 | | Crystal structure of TPADO in complex with 2-OH-TPA | | Descriptor: | 2-Hydroxyterephthalic acid, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q05
 
 | | Crystal structure of TPADO in complex with TPA | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Lysozyme, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q04
 
 | | Crystal structure of TPADO in a substrate-free state | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Lysozyme, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3OTP
 
 | |
1DQJ
 
 | |
3M18
 
 | |
3ULR
 
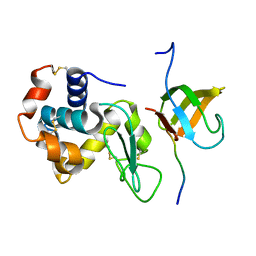 | | Lysozyme contamination facilitates crystallization of a hetero-trimericCortactin:Arg:Lysozyme complex | | Descriptor: | Abelson tyrosine-protein kinase 2, Lysozyme C, Src substrate cortactin | | Authors: | Liu, W, MacGrath, S, Koleske, A.J, Boggon, T.J. | | Deposit date: | 2011-11-11 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Lysozyme contamination facilitates crystallization of a heterotrimeric cortactin-Arg-lysozyme complex.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1J1O
 
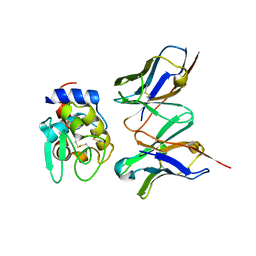 | | Crystal Structure of HyHEL-10 Fv mutant LY50F complexed with hen egg white lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Yokota, A, Tsumoto, K, Shiroishi, M, Kondo, H, Kumagai, I. | | Deposit date: | 2002-12-13 | | Release date: | 2003-01-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Role of Hydrogen Bonding via Interfacial Water Molecules in Antigen-Antibody Complexation. THE HyHEL-10-HEL INTERACTION
J.BIOL.CHEM., 278, 2003
|
|
1J1X
 
 | | Crystal Structure of HyHEL-10 Fv mutant LS93A complexed with hen egg white lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Yokota, A, Tsumoto, K, Shiroishi, M, Kondo, H, Kumagai, I. | | Deposit date: | 2002-12-20 | | Release date: | 2003-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Role of Hydrogen Bonding via Interfacial Water Molecules in Antigen-Antibody Complexation. THE HyHEL-10-HEL INTERACTION
J.BIOL.CHEM., 278, 2003
|
|
2I25
 
 | |
2I26
 
 | |
5JEN
 
 | |
4CJ2
 
 | | Crystal structure of HEWL in complex with affitin H4 | | Descriptor: | AFFITIN H4, GLYCEROL, LYSOZYME C | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
4A8A
 
 | | Asymmetric cryo-EM reconstruction of E. coli DegQ 12-mer in complex with lysozyme | | Descriptor: | LYSOZYME C, PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14.2 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
