1SAK
 
 | |
1SAE
 
 | |
1W25
 
 | | Response regulator PleD in complex with c-diGMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, STALKED-CELL DIFFERENTIATION CONTROLLING PROTEIN, ... | | Authors: | Chan, C, Schirmer, T, Jenal, U. | | Deposit date: | 2004-06-28 | | Release date: | 2004-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Activity and Allosteric Control of Diguanylate Cyclase
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1D2S
 
 | | CRYSTAL STRUCTURE OF THE N-TERMINAL LAMININ G-LIKE DOMAIN OF SHBG IN COMPLEX WITH DIHYDROTESTOSTERONE | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, CALCIUM ION, SEX HORMONE-BINDING GLOBULIN | | Authors: | Grishkovskaya, I, Avvakumov, G.V, Sklenar, G, Dales, D, Hammond, G.L, Muller, Y.A. | | Deposit date: | 1999-09-28 | | Release date: | 2000-06-28 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of human sex hormone-binding globulin: steroid transport by a laminin G-like domain.
EMBO J., 19, 2000
|
|
1VPF
 
 | | STRUCTURE OF HUMAN VASCULAR ENDOTHELIAL GROWTH FACTOR | | Descriptor: | VASCULAR ENDOTHELIAL GROWTH FACTOR | | Authors: | Muller, Y.A, De Vos, A.M. | | Deposit date: | 1997-04-08 | | Release date: | 1998-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Vascular endothelial growth factor: crystal structure and functional mapping of the kinase domain receptor binding site.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
2QBX
 
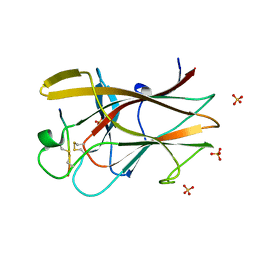 | | EphB2/SNEW Antagonistic Peptide Complex | | Descriptor: | Ephrin type-B receptor 2, SULFATE ION, antagonistic peptide | | Authors: | Chrencik, J.E, Brooun, A, Recht, M.I, Nicola, G, Pasquale, E.B, Kuhn, P, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-06-18 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the EphB2 receptor in complex with an antagonistic peptide reveals a novel mode of inhibition.
J.Biol.Chem., 282, 2007
|
|
1SPI
 
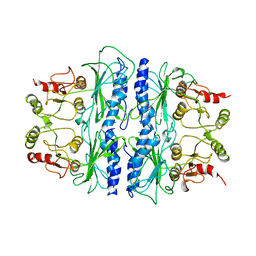 | | CRYSTAL STRUCTURE OF SPINACH CHLOROPLAST FRUCTOSE-1,6-BISPHOSPHATASE AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Villeret, V, Huang, S, Zhang, Y, Xue, Y, Lipscomb, W.N. | | Deposit date: | 1994-12-14 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of spinach chloroplast fructose-1,6-bisphosphatase at 2.8 A resolution.
Biochemistry, 34, 1995
|
|
1SAF
 
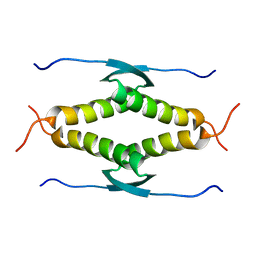 | |
1TCA
 
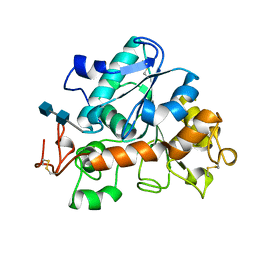 | |
1TLH
 
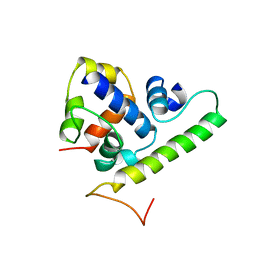 | | T4 AsiA bound to sigma70 region 4 | | Descriptor: | 10 kDa anti-sigma factor, RNA polymerase sigma factor rpoD | | Authors: | Lambert, L.J, Wei, Y, Schirf, V, Demeler, B, Werner, M.H. | | Deposit date: | 2004-06-09 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | T4 AsiA blocks DNA recognition by remodeling sigma(70) region 4
Embo J., 23, 2004
|
|
1T1N
 
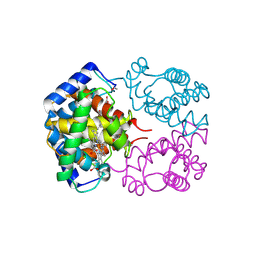 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN | | Descriptor: | CARBON MONOXIDE, PROTEIN (HEMOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mazzarella, L, Vitagliano, L, Savino, C, Zagari, A. | | Deposit date: | 1999-03-05 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Trematomus newnesi haemoglobin re-opens the root effect question.
J.Mol.Biol., 287, 1999
|
|
2QL1
 
 | | Structural Characterization of a Mutated, ADCC-Enhanced Human Fc Fragment | | Descriptor: | IGHM protein, ZINC ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Oganesyan, V, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2007-07-12 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.534 Å) | | Cite: | Structural characterization of a mutated, ADCC-enhanced human Fc fragment
Mol.Immunol., 45, 2008
|
|
1CLP
 
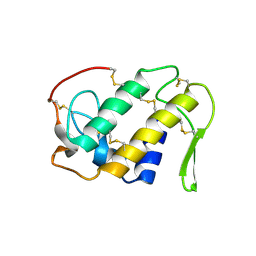 | | CRYSTAL STRUCTURE OF A CALCIUM-INDEPENDENT PHOSPHOLIPASELIKE MYOTOXIC PROTEIN FROM BOTHROPS ASPER VENOM | | Descriptor: | MYOTOXIN II | | Authors: | Arni, R.K, Ward, R.J, Gutierrez, J.M, Tulinsky, A. | | Deposit date: | 1994-09-12 | | Release date: | 1994-11-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a calcium-independent phospholipase-like myotoxic protein from Bothrops asper venom.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
2QSQ
 
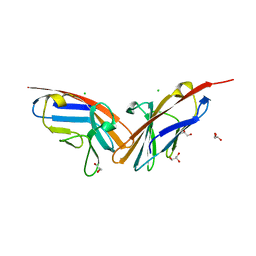 | | Crystal structure of the N-terminal domain of carcinoembryonic antigen (CEA) | | Descriptor: | CHLORIDE ION, Carcinoembryonic antigen-related cell adhesion molecule 5, GLYCEROL | | Authors: | Le Trong, I, Korotkova, N, Moseley, S.L, Stenkamp, R.E. | | Deposit date: | 2007-07-31 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Binding of Dr adhesins of Escherichia coli to carcinoembryonic antigen triggers receptor dissociation.
Mol.Microbiol., 67, 2008
|
|
2R31
 
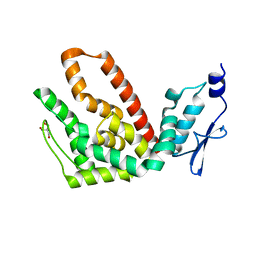 | | Crystal structure of atp12p from paracoccus denitrificans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP12 ATPase | | Authors: | Ludlam, A.V, Brunzelle, J.S, Gatti, D.L, Ackerman, S.H. | | Deposit date: | 2007-08-28 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Chaperones of F1-ATPase.
J.Biol.Chem., 284, 2009
|
|
2ARA
 
 | |
1D6X
 
 | |
1TAR
 
 | |
2NOF
 
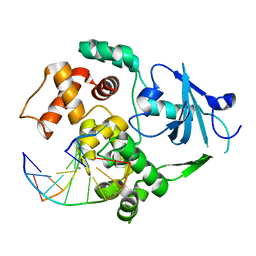 | | Structure of Q315F human 8-oxoguanine glycosylase proximal crosslink to 8-oxoguanine DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*C*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOI
 
 | | Structure of G42A human 8-oxoguanine glycosylase crosslinked to undamaged G-containing DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*C*CP*AP*GP*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
1DAF
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, ADP, AND CALCIUM | | Descriptor: | 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
1DAE
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 3-(1-AMINOETHYL) NONANEDIOIC ACID | | Descriptor: | 3-(1-AMINOETHYL)NONANEDIOIC ACID, DETHIOBIOTIN SYNTHETASE | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
2NOB
 
 | | Structure of catalytically inactive H270A human 8-oxoguanine glycosylase crosslinked to 8-oxoguanine DNA | | Descriptor: | 5'-D(*G*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*T*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
1DHM
 
 | | DNA-BINDING DOMAIN OF E2 FROM HUMAN PAPILLOMAVIRUS-31, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | E2 PROTEIN | | Authors: | Liang, H, Petros, A.P, Meadows, R.P, Yoon, H.S, Egan, D.A, Walter, K, Holzman, T.F, Robins, T, Fesik, S.W. | | Deposit date: | 1995-08-15 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of a human papillomavirus E2 protein: evidence for flexible DNA-binding regions.
Biochemistry, 35, 1996
|
|
2B4L
 
 | | Crystal structure of the binding protein OpuAC in complex with glycine betaine | | Descriptor: | 1,2-ETHANEDIOL, Glycine betaine-binding protein, TRIMETHYL GLYCINE | | Authors: | Horn, C, Sohn-Boesser, L, Breed, J, Welte, W, Schmitt, L, Bremer, E. | | Deposit date: | 2005-09-26 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Determinants for Substrate Specificity of the Ligand-binding Protein OpuAC from Bacillus subtilis for the Compatible Solutes Glycine Betaine and Proline Betaine.
J.Mol.Biol., 357, 2006
|
|
