1BIZ
 
 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-21 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
5F8J
 
 | | Enterovirus 71 Polymerase Elongation Complex (C1S4 Form) | | Descriptor: | GLYCEROL, Genome polyprotein, MAGNESIUM ION, ... | | Authors: | Shu, B, Gong, P. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.675 Å) | | Cite: | Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8DID
 
 | |
1C0R
 
 | | COMPLEX OF VANCOMYCIN WITH D-LACTIC ACID | | Descriptor: | CHLORIDE ION, LACTIC ACID, VANCOMYCIN, ... | | Authors: | Loll, P.J, Kaplan, J, Selinsky, B, Axelsen, P.H. | | Deposit date: | 1999-07-20 | | Release date: | 1999-07-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Vancomycin Binding to Low-Affinity Ligands: Delineating a Minimum Set of Interactions Necessary for High-Affinity Binding.
J.Med.Chem., 42, 1999
|
|
1C24
 
 | | E. COLI METHIONINE AMINOPEPTIDASE: METHIONINE PHOSPHINATE COMPLEX | | Descriptor: | (1-AMINO-3-METHYLSULFANYL-PROPYL)-PHOSPHINIC ACID, COBALT (II) ION, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Lowther, W.T, Zhang, Y, Sampson, P.B, Honek, J.F, Matthews, B.W. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the mechanism of Escherichia coli methionine aminopeptidase from the structural analysis of reaction products and phosphorus-based transition-state analogues.
Biochemistry, 38, 1999
|
|
6HIJ
 
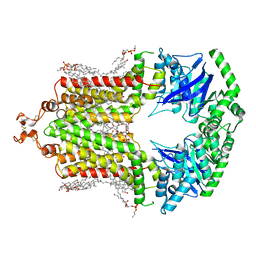 | | Cryo-EM structure of the human ABCG2-MZ29-Fab complex with cholesterol and PE lipids docked | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ATP-binding cassette sub-family G member 2, CHOLESTEROL, ... | | Authors: | Jackson, S.M, Manolaridis, I, Kowal, J, Zechner, M, Taylor, N.M.I, Bause, M, Bauer, S, Bartholomaeus, R, Stahlberg, H, Bernhardt, G, Koenig, B, Buschauer, A, Altmann, K.H, Locher, K.P. | | Deposit date: | 2018-08-30 | | Release date: | 2018-09-19 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Nat.Struct.Mol.Biol., 25, 2018
|
|
5ACO
 
 | | Cryo-EM structure of PGT128 Fab in complex with BG505 SOSIP.664 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 ENVELOPE GLYCOPROTEIN, ... | | Authors: | Lee, J.H, Ward, A.B. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-30 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Model Building and Refinement of a Natively Glycosylated HIV-1 Env Protein by High-Resolution Cryoelectron Microscopy.
Structure, 23, 2015
|
|
5LVT
 
 | |
8FUH
 
 | |
5LY0
 
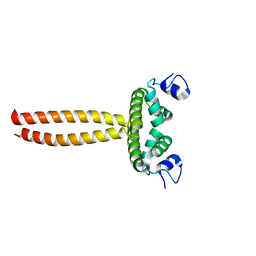 | | Crystal structure of LOB domain of Ramosa2 from Wheat | | Descriptor: | LOB family transfactor Ramosa2.1, ZINC ION | | Authors: | Wei, X.-B, Zhang, B, Chen, W.-F, Fan, S.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-23 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Structural analysis reveals a "molecular calipers" mechanism for a LATERAL ORGAN BOUNDARIES DOMAIN transcription factor protein from wheat.
J. Biol. Chem., 294, 2019
|
|
6SZU
 
 | | Bat Influenza A polymerase pre-termination complex with pyrophosphate using 44-mer vRNA template with mutated oligo(U) sequence | | Descriptor: | 5-oxidanyl-4-oxidanylidene-1-[(1-pyrrolo[2,3-b]pyridin-1-ylcyclopentyl)methyl]pyridine-3-carboxylic acid, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Wandzik, J.M, Kouba, T, Cusack, S. | | Deposit date: | 2019-10-02 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
4XHB
 
 | |
4XJW
 
 | |
5LTN
 
 | | Crystal structure of Lymphocytic choriomeningitis mammarenavirus endonuclease complexed with DPBA | | Descriptor: | 2-4-DIOXO-4-PHENYLBUTANOIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saez-Ayala, M, Yekwa, E.L, Canard, B, Alvarez, K, Ferron, F. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of Lymphocytic choriomeningitis virusendonuclease domain complexed with diketo-acid ligands.
Iucrj, 5, 2018
|
|
8FVV
 
 | |
4XD0
 
 | | X-ray structure of the N-formyltransferase QdtF from Providencia alcalifaciens | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, CHLORIDE ION, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Thoden, J.B, Woodford, C.R, Holden, H.M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New Role for the Ankyrin Repeat Revealed by a Study of the N-Formyltransferase from Providencia alcalifaciens.
Biochemistry, 54, 2015
|
|
8FXD
 
 | |
1AUA
 
 | | PHOSPHATIDYLINOSITOL TRANSFER PROTEIN SEC14P FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | PHOSPHATIDYLINOSITOL TRANSFER PROTEIN SEC14P, octyl beta-D-glucopyranoside | | Authors: | Sha, B, Phillips, S.E, Bankaitis, V.A, Luo, M. | | Deposit date: | 1997-08-20 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein.
Nature, 391, 1998
|
|
6SNV
 
 | | DNA mismatch repair proteins MLH1 and MLH3 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein MLH3, ZINC ION | | Authors: | Dai, J, Chervy, P, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2019-08-27 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5FIC
 
 | | Open form of murine Acid Sphingomyelinase in presence of lipid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Heinz, L.X, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of mammalian acid sphingomyelinase.
Nat Commun, 7, 2016
|
|
8G1M
 
 | |
8BO1
 
 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, bound to Latrunculin-B-ATP-Mg-actin, and 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE and 2 Mg ions | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AZIDE ION, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
4XJ9
 
 | |
8G1L
 
 | |
4XJZ
 
 | |
