7K4C
 
 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor Br-cis-22a | | Descriptor: | 1-(5-bromopyridin-3-yl)-4-[cis-4-(3-methylphenyl)cyclohexyl]piperazine, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7L2H
 
 | | Cryo-EM structure of unliganded full-length TRPV1 at neutral pH | | Descriptor: | (10R,13S)-16-amino-13-hydroxy-7,13-dioxo-8,12,14-trioxa-13lambda~5~-phosphahexadecan-10-yl tridecanoate, (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2O
 
 | |
7L2J
 
 | | Cryo-EM structure of full-length TRPV1 at pH6c state | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2I
 
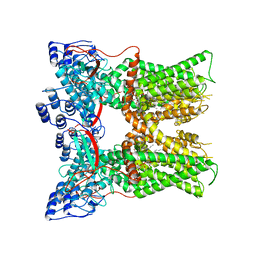 | | Cryo-EM structure of full-length TRPV1 at pH6a state | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1S,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2K
 
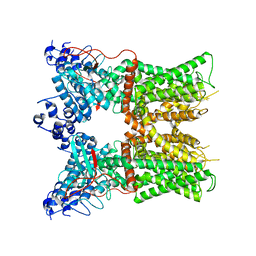 | |
7L2M
 
 | | Cryo-EM structure of DkTx/RTX-bound full-length TRPV1 | | Descriptor: | SODIUM ION, Tau-theraphotoxin-Hs1a, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2N
 
 | |
7L2L
 
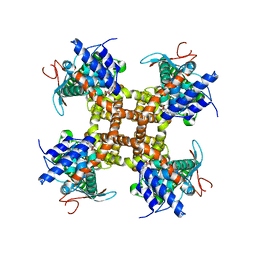 | |
2B1O
 
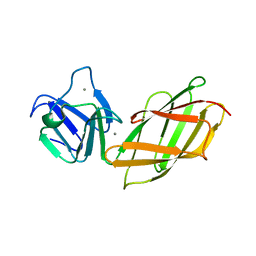 | | Solution Structure of Ca2+-bound DdCAD-1 | | Descriptor: | CALCIUM ION, Calcium-dependent cell adhesion molecule-1 | | Authors: | Lin, Z, Sriskanthadevan, S, Huang, H.B, Siu, C.H, Yang, D.W. | | Deposit date: | 2005-09-16 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the adhesion molecule DdCAD-1 reveal new insights into Ca(2+)-dependent cell-cell adhesion
Nat.Struct.Mol.Biol., 13, 2006
|
|
2AQX
 
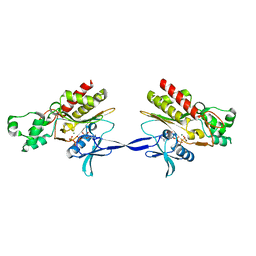 | | Crystal Structure of the Catalytic and CaM-Binding domains of Inositol 1,4,5-Trisphosphate 3-Kinase B | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PREDICTED: inositol 1,4,5-trisphosphate 3-kinase B | | Authors: | Chamberlain, P.P, Sandberg, M.L, Sauer, K, Cooke, M.P, Lesley, S.A, Spraggon, G. | | Deposit date: | 2005-08-18 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into enzyme regulation for inositol 1,4,5-trisphosphate 3-kinase B
Biochemistry, 44, 2005
|
|
2C32
 
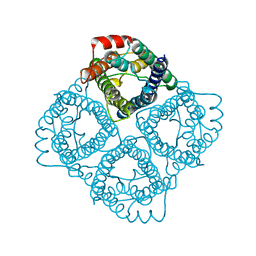 | |
2BL0
 
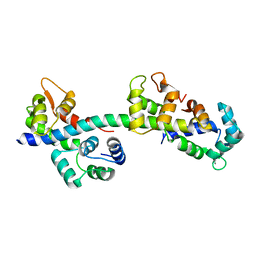 | | Physarum polycephalum myosin II regulatory domain | | Descriptor: | CALCIUM ION, MAJOR PLASMODIAL MYOSIN HEAVY CHAIN, MYOSIN REGULATORY LIGHT CHAIN | | Authors: | Debreczeni, J.E, Farkas, L, Harmat, V, Nyitray, L. | | Deposit date: | 2005-02-23 | | Release date: | 2005-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Evidence for Non-Canonical Binding of Ca2+ to a Canonical EF-Hand of a Conventional Myosin.
J.Biol.Chem., 280, 2005
|
|
2B6O
 
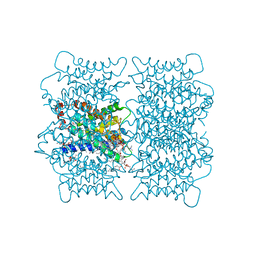 | | Electron crystallographic structure of lens Aquaporin-0 (AQP0) (lens MIP) at 1.9A resolution, in a closed pore state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Lens fiber major intrinsic protein | | Authors: | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | Deposit date: | 2005-10-03 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | Cite: | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
2B6P
 
 | | X-ray structure of lens Aquaporin-0 (AQP0) (lens MIP) in an open pore state | | Descriptor: | Lens fiber major intrinsic protein | | Authors: | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | Deposit date: | 2005-10-03 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
2D21
 
 | | NMR Structure of stereo-array isotope labelled (SAIL) maltodextrin-binding protein (MBP) | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Kainosho, M, Torizawa, T, Iwashita, Y, Terauchi, T, Ono, A.M, Guntert, P. | | Deposit date: | 2005-09-02 | | Release date: | 2006-03-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Optimal isotope labelling for NMR protein structure determinations.
Nature, 440, 2006
|
|
8DVV
 
 | | Recombinant mouse RyR2 triple phosphomimetic mutant S2807D/S2813D/S2030D in complex with FKBP12.6 and nanodisc under open-state conditions | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Iyer, K.A, Hu, Y, Murayama, T, Samso, M. | | Deposit date: | 2022-07-29 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Recombinant mouse RyR2 triple phosphomimetic mutant S2807D/S2813D/S2030D in complex with FKBP12.6 and nanodisc under open-state conditions
To Be Published
|
|
8DTY
 
 | |
8DTZ
 
 | |
1PNB
 
 | | STRUCTURE OF NAPIN BNIB, NMR, 10 STRUCTURES | | Descriptor: | NAPIN BNIB | | Authors: | Rico, M, Bruix, M, Gonzalez, C, Monsalve, R, Rodriguez, R. | | Deposit date: | 1996-09-17 | | Release date: | 1997-09-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | 1H NMR assignment and global fold of napin BnIb, a representative 2S albumin seed protein.
Biochemistry, 35, 1996
|
|
7VUS
 
 | | Crystal structure of AlleyCat9 with 5-nitro-benzotriazole | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-nitro-1H-benzotriazole, AlleyCat, ... | | Authors: | Tame, J.R.H, Korendovych, I.V, Margheritis, E, Takahashi, K. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
7VUR
 
 | | Crystal structure of AlleyCat9 with calcium but no inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, AlleyCat, CALCIUM ION | | Authors: | Margheritis, E, Takahashi, K, Korendovych, I.V, Tame, J.R.H. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
1QLK
 
 | | SOLUTION STRUCTURE OF CA(2+)-LOADED RAT S100B (BETABETA) NMR, 20 STRUCTURES | | Descriptor: | CALCIUM ION, S-100 PROTEIN | | Authors: | Drohat, A.C, Baldisseri, D.M, Rustandi, R.R, Weber, D.J. | | Deposit date: | 1997-09-26 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calcium-bound rat S100B(betabeta) as determined by nuclear magnetic resonance spectroscopy,.
Biochemistry, 37, 1998
|
|
1P4F
 
 | | DEATH ASSOCIATED PROTEIN KINASE CATALYTIC DOMAIN WITH BOUND INHIBITOR FRAGMENT | | Descriptor: | 5,6-Dihydro-benzo[H]cinnolin-3-ylamine, Death-associated protein kinase 1 | | Authors: | Velentza, A.V, Wainwright, M.S, Zasadzki, M, Mirzoeva, S, Haiech, J, Focia, P.J, Egli, M, Watterson, D.M. | | Deposit date: | 2003-04-23 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An aminopyridazine-based inhibitor of a pro-apoptotic protein kinase attenuates hypoxia-ischemia induced acute brain injury.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
8EMX
 
 | | Phospholipase C beta 3 (PLCb3) in complex with Gbg on lipid nanodiscs | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | G beta gamma activates PIP2 hydrolysis by recruiting and orienting PLC beta on the membrane surface.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
