2LFM
 
 | |
2BKG
 
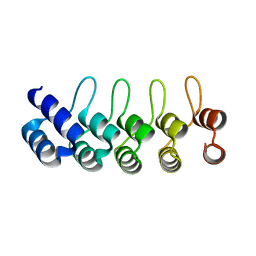 | | Crystal structure of E3_19 a designed ankyrin repeat protein | | Descriptor: | SYNTHETIC CONSTRUCT ANKYRIN REPEAT PROTEIN E3_19 | | Authors: | Binz, H.K, Kohl, A, Pluckthun, A, Grutter, M.G. | | Deposit date: | 2005-02-16 | | Release date: | 2006-06-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Consensus-Designed Ankyrin Repeat Protein: Implications for Stability
Proteins: Struct., Funct., Bioinf., 65, 2006
|
|
2L0P
 
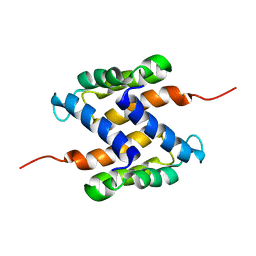 | | Solution structure of human apo-S100A1 protein by NMR spectroscopy | | Descriptor: | S100 calcium binding protein A1 | | Authors: | Nowakowski, M, Jaremko, L, Jaremko, M, Bierzynski, A, Zhukov, I, Ejchart, A. | | Deposit date: | 2010-07-12 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and dynamics of human apo-S100A1 protein.
J.Struct.Biol., 174, 2011
|
|
4E1B
 
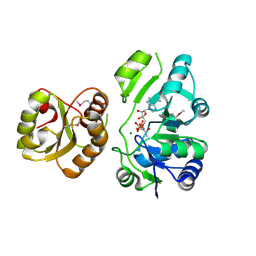 | | Re-refinement of PDB entry 2EQA - SUA5 protein from Sulfolobus tokodaii with bound threonylcarbamoyladenylate | | Descriptor: | MAGNESIUM ION, YrdC/Sua5 family protein, threonylcarbamoyladenylate | | Authors: | Parthier, C, Goerlich, S, Jaenecke, F, Breithaupt, C, Braeuer, U, Fandrich, U, Clausnitzer, D, Wehmeier, U.F, Boettcher, C, Scheel, D, Stubbs, M.T. | | Deposit date: | 2012-03-06 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
2Z29
 
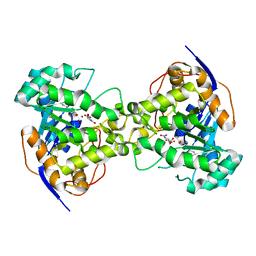 | | Thr109Ala dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
4DPE
 
 | | Structure of MMP3 complexed with a platinum-based inhibitor. | | Descriptor: | CALCIUM ION, CHLORIDE ION, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Belviso, B.D, Arnesano, F, Calderone, V, Caliandro, R, Natile, G, Siliqi, D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of matrix metalloproteinase-3 with a platinum-based inhibitor.
Chem.Commun.(Camb.), 49, 2013
|
|
2KXW
 
 | |
2KMU
 
 | |
2LWP
 
 | |
2MNI
 
 | |
2MEM
 
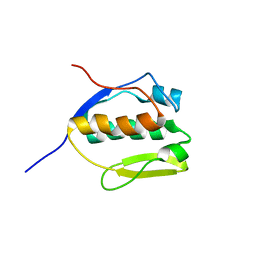 | | Solution NMR structure of SLED domain of Scml2 | | Descriptor: | Sex comb on midleg-like protein 2 | | Authors: | Bezsonova, I. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the DNA-binding Domain from Scml2 (Sex Comb on Midleg-like 2).
J.Biol.Chem., 289, 2014
|
|
2LXZ
 
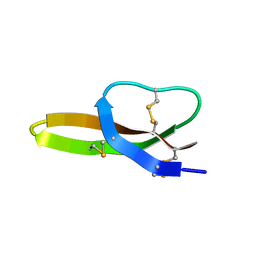 | | Solution Structure of the Antimicrobial Peptide Human Defensin 5 | | Descriptor: | Defensin-5 | | Authors: | Wommack, A.J, Robson, S.A, Wanniarahchi, Y.A, Wan, A, Turner, C.J, Nolan, E.M. | | Deposit date: | 2012-09-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and condition-dependent oligomerization of the antimicrobial Peptide human defensin 5.
Biochemistry, 51, 2012
|
|
2MUH
 
 | |
2HZP
 
 | | Crystal Structure of Homo Sapiens Kynureninase | | Descriptor: | Kynureninase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lima, S, Khristoforov, R, Momany, C, Phillips, R.S. | | Deposit date: | 2006-08-09 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Homo sapiens Kynureninase.
Biochemistry, 46, 2007
|
|
2PZ0
 
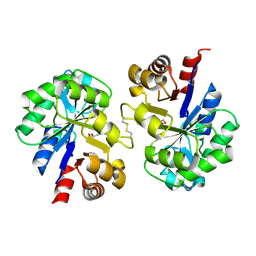 | | Crystal structure of Glycerophosphodiester Phosphodiesterase (GDPD) from T. tengcongensis | | Descriptor: | CALCIUM ION, GLYCEROL, Glycerophosphoryl diester phosphodiesterase | | Authors: | Shi, L, Liu, J.F, An, X.M, Liang, D.C. | | Deposit date: | 2007-05-17 | | Release date: | 2008-04-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of glycerophosphodiester phosphodiesterase (GDPD) from Thermoanaerobacter tengcongensis, a metal ion-dependent enzyme: insight into the catalytic mechanism.
Proteins, 72, 2008
|
|
4C2J
 
 | | Crystal structure of human mitochondrial 3-ketoacyl-CoA thiolase in complex with CoA | | Descriptor: | 1,2-ETHANEDIOL, 3-KETOACYL-COA THIOLASE, MITOCHONDRIAL, ... | | Authors: | Kiema, T.-R, Harijan, R.K, Wierenga, R.K. | | Deposit date: | 2013-08-19 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Mitochondrial 3-Ketoacyl-Coa Thiolase (T1): Insight Into the Reaction Mechanism of its Thiolase and Thioesterase Activities
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2IHN
 
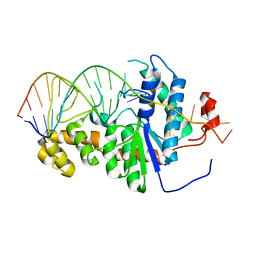 | | Co-crystal of Bacteriophage T4 RNase H with a fork DNA substrate | | Descriptor: | 5'-D(*CP*TP*AP*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*CP*C)-3', 5'-D(*GP*GP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*TP*AP*GP*TP*CP*AP*A)-3', Ribonuclease H | | Authors: | Devos, J.M, Mueser, T.C. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of bacteriophage T4 5' nuclease in complex with a branched DNA reveals how FEN-1 family nucleases bind their substrates.
J.Biol.Chem., 282, 2007
|
|
1YMQ
 
 | |
4A3U
 
 | |
3PRC
 
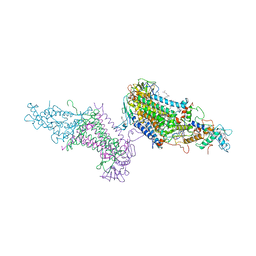 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (QB-DEPLETED) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-07-29 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The coupling of light-induced electron transfer and proton uptake as derived from crystal structures of reaction centres from Rhodopseudomonas viridis modified at the binding site of the secondary quinone, QB.
Structure, 5, 1997
|
|
3UNX
 
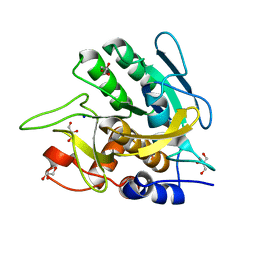 | | Bond length analysis of asp, glu and his residues in subtilisin Carlsberg at 1.26A resolution | | Descriptor: | CALCIUM ION, GLYCEROL, SODIUM ION, ... | | Authors: | Fisher, S.J, Helliwell, J.R, Blakeley, M.P, Cianci, M, McSweeny, S. | | Deposit date: | 2011-11-16 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Protonation-state determination in proteins using high-resolution X-ray crystallography: effects of resolution and completeness.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2CHR
 
 | |
2KVJ
 
 | | NMR and MD solution structure of a Gamma-Methylated PNA duplex | | Descriptor: | Gamma-Modified Peptide Nucleic Acid | | Authors: | He, W, Crawford, M.J, Rapireddy, S, Madrid, M, Gil, R.R, Ly, D.H, Achim, C. | | Deposit date: | 2010-03-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The structure of a gamma-modified peptide nucleic acid duplex.
Mol Biosyst, 6, 2010
|
|
2CWQ
 
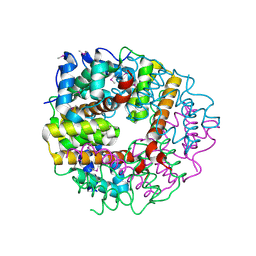 | | Crystal structure of conserved protein TTHA0727 from Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TTHA0727 | | Authors: | Ito, K, Arai, R, Fusatomi, E, Kamo-Uchikubo, T, Kawaguchi, S, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-23 | | Release date: | 2005-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the conserved protein TTHA0727 from Thermus thermophilus HB8 at 1.9 A resolution: A CMD family member distinct from carboxymuconolactone decarboxylase (CMD) and AhpD
Protein Sci., 15, 2006
|
|
2OTC
 
 | |
