1R3X
 
 | | INTRAMOLECULAR DNA TRIPLEX WITH RNA THIRD STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*CP*T)-3'), RNA (5'-R(*UP*CP*UP*CP*UP*CP*UP*U)-3') | | Authors: | Gotfredsen, C.H, Schultze, P, Feigon, J. | | Deposit date: | 1998-02-06 | | Release date: | 1998-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of an Intramolecular Pyrimidine-Purine-Pyrimidine Triplex Containing an RNA Third Strand
J.Am.Chem.Soc., 120, 1998
|
|
7YS7
 
 | | RET oncogene primer G4-DNA in 100mMNa+ | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3') | | Authors: | Yin, S, Cao, C. | | Deposit date: | 2022-08-11 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RET oncogene primer G-quadruplex in Na+
To Be Published
|
|
8SJV
 
 | | [4T24] Self-assembling left-handed tensegrity triangle with 24 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(P*CP*TP*TP*GP*TP*AP*GP*TP*CP*TP*CP*AP*CP*CP*AP*CP*TP*GP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*GP*AP*AP*CP*AP*CP*TP*CP*CP*TP*GP*AP*GP*AP*CP*TP*AP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*CP*AP*TP*CP*AP*CP*AP*GP*TP*GP*GP*AP*CP*TP*AP*CP*AP*AP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (8.59 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJR
 
 | | [3T17] Self-assembling right-handed tensegrity triangle with 17 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*GP*CP*CP*TP*GP*AP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*CP*TP*GP*TP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*GP*TP*AP*TP*TP*CP*AP*CP*CP*AP*CP*GP*AP*T)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (5.25 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJM
 
 | | [3T12] Self-assembling left-handed tensegrity triangle with 12 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*CP*GP*CP*CP*TP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*GP*CP*AP*TP*GP*TP*GP*GP*CP*GP*AP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*AP*TP*GP*CP*GP*AP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (8.08 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJQ
 
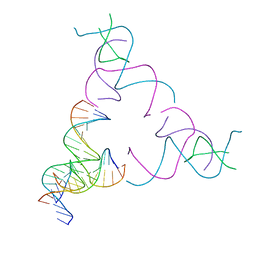 | | [3T16] Self-assembling right-handed tensegrity triangle with 16 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*GP*CP*CP*TP*GP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*CP*TP*GP*TP*GP*GP*CP*AP*TP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*TP*GP*GP*AP*CP*AP*GP*CP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (6.19 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJN
 
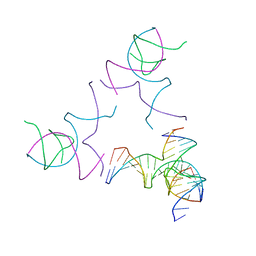 | | [3T13] Self-assembling left-handed tensegrity triangle with 13 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*CP*GP*CP*CP*TP*GP*AP*CP*TP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*GP*CP*TP*GP*TP*GP*GP*CP*GP*AP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*AP*GP*CP*GP*AP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7.3 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJS
 
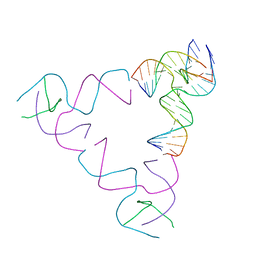 | | [3T18] Self-assembling right-handed tensegrity triangle with 18 interjunction base pairs and P63 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*GP*CP*CP*TP*GP*AP*CP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*CP*TP*GP*TP*GP*GP*CP*TP*C)-3'), DNA (5'-D(P*TP*CP*GP*TP*GP*GP*AP*CP*AP*GP*CP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (6.31 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJW
 
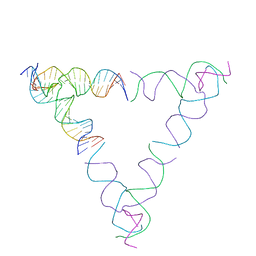 | | [4T28] Self-assembling right-handed tensegrity triangle with 28 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (28-MER), DNA (5'-D(*GP*AP*AP*CP*TP*GP*CP*CP*TP*GP*AP*AP*TP*TP*AP*CP*TP*GP*AP*CP*CP*G)-3'), DNA (5'-D(*TP*CP*AP*TP*CP*AP*GP*TP*GP*GP*CP*AP*GP*T)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7.64 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJU
 
 | | [4T17] Self-assembling right-handed four-turn tensegrity triangle with 17 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (25-MER), DNA (5'-D(*GP*AP*AP*AP*AP*AP*CP*AP*CP*TP*GP*CP*CP*TP*GP*AP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(P*GP*CP*GP*GP*TP*AP*TP*TP*CP*AP*CP*CP*AP*CP*GP*AP*T)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7.14 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJO
 
 | | [3T14] Self-assembling left-handed tensegrity triangle with 14 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(P*CP*AP*CP*GP*TP*GP*GP*AP*CP*AP*GP*GP*AP*G)-3'), DNA (5'-D(P*CP*AP*GP*CP*TP*CP*AP*GP*CP*CP*TP*GP*AP*CP*TP*CP*A)-3'), DNA (5'-D(P*GP*TP*GP*AP*GP*TP*CP*TP*CP*CP*AP*CP*GP*T)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7.08 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SL5
 
 | | [2T13] Self-assembling left-handed two-turn tensegrity triangle with 13 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*CP*TP*GP*AP*CP*TP*AP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*TP*GP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*A)-3'), ... | | Authors: | Vecchioni, S, Janowski, J, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (6.55 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3R7P
 
 | | The crystal structure of I-LtrI | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*TP*GP*CP*TP*CP*CP*TP*AP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*CP*TP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(P*AP*GP*GP*AP*GP*CP*AP*TP*TP*TP*G)-3'), ... | | Authors: | Mak, A.N.S, Takeuchi, R, Edgell, D.R, Stoddard, B.L. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Tapping natural reservoirs of homing endonucleases for targeted gene modification.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7CRQ
 
 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (2:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRO
 
 | | NSD2 bearing E1099K/T1150A dual mutation in complex with 187-bp NCP | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7V6E
 
 | | DREP3 | | Descriptor: | DNAation factor-related protein 3, isoform A | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Helical filament structure of the DREP3 CIDE domain reveals a unified mechanism of CIDE-domain assembly.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8X1V
 
 | |
8I4W
 
 | | Cryo-EM structure of 5-subunit Smc5/6 head region | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 5, Structural maintenance of chromosomes protein 5, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Cheng, T, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.01 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8I4V
 
 | | Cryo-EM structure of 5-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.97 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
7CRP
 
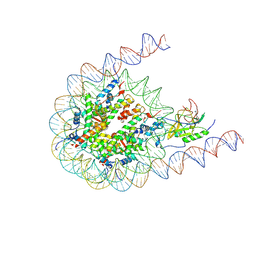 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (1:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
2B2Y
 
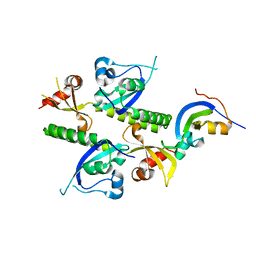 | | Tandem chromodomains of human CHD1 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
2B2U
 
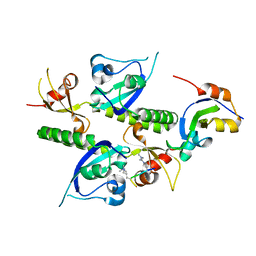 | | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 and dimethylarginine 2 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
2B2V
 
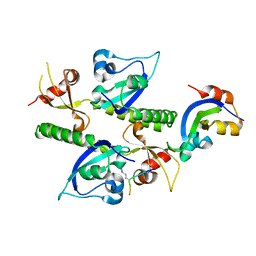 | | Crystal structure analysis of human CHD1 chromodomains 1 and 2 bound to histone H3 resi 1-15 MeK4 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
2L5U
 
 | |
6PNK
 
 | |
