1NK3
 
 | | VND/NK-2 HOMEODOMAIN/DNA COMPLEX, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*CP*TP*GP*T)-3'), HOMEOBOX PROTEIN VND | | Authors: | Gruschus, J.M, Tsao, D.H.H, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1998-05-06 | | Release date: | 1998-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity.
Biochemistry, 36, 1997
|
|
1NK2
 
 | | VND/NK-2 HOMEODOMAIN/DNA COMPLEX, NMR, 20 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*CP*TP*GP*T)-3'), HOMEOBOX PROTEIN VND | | Authors: | Gruschus, J.M, Tsao, D.H.H, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1998-05-06 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity.
Biochemistry, 36, 1997
|
|
1Y8Y
 
 | | Crystal structure of human CDK2 complexed with a pyrazolo[1,5-a]pyrimidine inhibitor | | Descriptor: | (5-CHLOROPYRAZOLO[1,5-A]PYRIMIDIN-7-YL)-(4-METHANESULFONYLPHENYL)AMINE, Cell division protein kinase 2 | | Authors: | Williamson, D.S, Parratt, M.J, Torrance, C.J, Bower, J.F, Moore, J.D, Richardson, C.M, Dokurno, P, Cansfield, A.D, Francis, G.L, Hebdon, R.J, Howes, R, Jackson, P.S, Lockie, A.M, Murray, J.B, Nunns, C.L, Powles, J, Robertson, A, Surgenor, A.E. | | Deposit date: | 2004-12-14 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Structure-guided design of pyrazolo[1,5-a]pyrimidines as inhibitors of human cyclin-dependent kinase 2.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1Y91
 
 | | Crystal structure of human CDK2 complexed with a pyrazolo[1,5-a]pyrimidine inhibitor | | Descriptor: | 4-[5-(TRANS-4-AMINOCYCLOHEXYLAMINO)-3-ISOPROPYLPYRAZOLO[1,5-A]PYRIMIDIN-7-YLAMINO]-N,N-DIMETHYLBENZENESULFONAMIDE, Cell division protein kinase 2 | | Authors: | Williamson, D.S, Parratt, M.J, Torrance, C.J, Bower, J.F, Moore, J.D, Richardson, C.M, Dokurno, P, Cansfield, A.D, Francis, G.L, Hebdon, R.J, Howes, R, Jackson, P.S, Lockie, A.M, Murray, J.B, Nunns, C.L, Powles, J, Robertson, A, Surgenor, A.E. | | Deposit date: | 2004-12-14 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-guided design of pyrazolo[1,5-a]pyrimidines as inhibitors of human cyclin-dependent kinase 2.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1R0E
 
 | | Glycogen synthase kinase-3 beta in complex with 3-indolyl-4-arylmaleimide inhibitor | | Descriptor: | 3-[3-(2,3-DIHYDROXY-PROPYLAMINO)-PHENYL]-4-(5-FLUORO-1-METHYL-1H-INDOL-3-YL)-PYRROLE-2,5-DIONE, CITRATE ANION, Glycogen synthase kinase-3 beta | | Authors: | Allard, J, Nikolcheva, T, Gong, L, Wang, J, Dunten, P, Avnur, Z, Waters, R, Sun, Q, Skinner, B. | | Deposit date: | 2003-09-20 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | From genetics to therapeutics: the Wnt pathway and osteoporosis
To be Published
|
|
1QMZ
 
 | | PHOSPHORYLATED CDK2-CYCLYIN A-SUBSTRATE PEPTIDE COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, G2/MITOTIC-SPECIFIC CYCLIN A, ... | | Authors: | Brown, N.R, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 1999-10-11 | | Release date: | 1999-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for Specificity of Substrate and Recruitment Peptides for Cyclin-Dependent Kinases
Nat.Cell Biol., 1, 1999
|
|
1QQI
 
 | | SOLUTION STRUCTURE OF THE DNA-BINDING AND TRANSACTIVATION DOMAIN OF PHOB FROM ESCHERICHIA COLI | | Descriptor: | PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | Authors: | Okamura, H, Hanaoka, S, Nagadoi, A, Makino, K, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-06-07 | | Release date: | 2000-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box.
J.Mol.Biol., 295, 2000
|
|
1YFD
 
 | | Crystal structure of the Y122H mutant of ribonucleotide reductase R2 protein from E. coli | | Descriptor: | MERCURY (II) ION, MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Kolberg, M, Logan, D.T, Bleifuss, G, Poetsch, S, Sjoeberg, B.M, Graeslund, A, Lubitz, W, Lassmann, G, Lendzian, F. | | Deposit date: | 2004-12-31 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new tyrosyl radical on Phe208 as ligand to the diiron center in Escherichia coli ribonucleotide reductase, mutant R2-Y122H. Combined x-ray diffraction and EPR/ENDOR studies
J.Biol.Chem., 280, 2005
|
|
1NYT
 
 | | SHIKIMATE DEHYDROGENASE AroE COMPLEXED WITH NADP+ | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Roszak, A.W, Lapthorn, A.J. | | Deposit date: | 2003-02-13 | | Release date: | 2003-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of shikimate dehydrogenase AroE and its Paralog YdiB. A common structural framework for different activities.
J.Biol.Chem., 278, 2003
|
|
1XGK
 
 | | CRYSTAL STRUCTURE OF N12G AND A18G MUTANT NMRA | | Descriptor: | CHLORIDE ION, GLYCEROL, NITROGEN METABOLITE REPRESSION REGULATOR NMRA, ... | | Authors: | Lamb, H.K, Ren, J, Park, A, Johnson, C, Leslie, K, Cocklin, S, Thompson, P, Mee, C, Cooper, A, Stammers, D.K, Hawkins, A.R. | | Deposit date: | 2004-09-17 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Modulation of the ligand binding properties of the transcription repressor NmrA by GATA-containing DNA and site-directed mutagenesis
Protein Sci., 13, 2004
|
|
1XKU
 
 | | Crystal structure of the dimeric protein core of decorin, the archetypal small leucine-rich repeat proteoglycan | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Decorin | | Authors: | Scott, P.G, McEwan, P.A, Dodd, C.M, Bergmann, E.M, Bishop, P.N, Bella, J. | | Deposit date: | 2004-09-29 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the dimeric protein core of decorin, the archetypal small leucine-rich repeat proteoglycan
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1Y7Q
 
 | | Mammalian SCAN domain dimer is a domain-swapped homologue of the HIV capsid C-terminal domain | | Descriptor: | Zinc finger protein 174 | | Authors: | Ivanov, D, Stone, J.R, Maki, J.L, Collins, T, Wagner, G. | | Deposit date: | 2004-12-09 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mammalian SCAN Domain Dimer Is a Domain-Swapped Homolog of the HIV Capsid C-Terminal Domain
Mol.Cell, 17, 2005
|
|
1B38
 
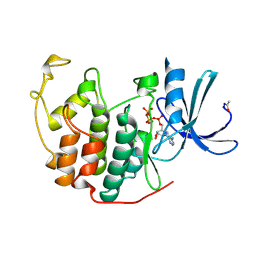 | | HUMAN CYCLIN-DEPENDENT KINASE 2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PROTEIN (CELL DIVISION PROTEIN KINASE 2) | | Authors: | Brown, N.R, Noble, M.E.M, Lawrie, A.M, Morris, M.C, Tunnah, P, Divita, G, Johnson, L.N, Endicott, J.A. | | Deposit date: | 1998-12-17 | | Release date: | 1998-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of phosphorylation of threonine 160 on cyclin-dependent kinase 2 structure and activity.
J.Biol.Chem., 274, 1999
|
|
1AX8
 
 | | Human obesity protein, leptin | | Descriptor: | OBESITY PROTEIN | | Authors: | Zhang, F, Beals, J.M, Briggs, S.L, Clawson, D.K, Wery, J.-P, Schevitz, R.W. | | Deposit date: | 1997-10-31 | | Release date: | 1998-11-25 | | Last modified: | 2012-05-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the obese protein leptin-E100.
Nature, 387, 1997
|
|
1AU7
 
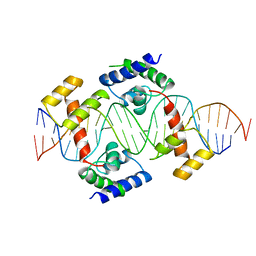 | | PIT-1 MUTANT/DNA COMPLEX | | Descriptor: | CONSENSUS DNA 25-MER, DNA (5'-D(*CP*TP*TP*CP*CP*TP*CP*AP*TP*GP*TP*AP*TP*AP*TP*AP*C P*AP*TP*GP*AP*GP* GP*A)-3'), PROTEIN PIT-1 | | Authors: | Jacobson, E.M, Li, P, Leon-Del-Rio, A, Rosenfeld, M.G, Aggarwal, A.K. | | Deposit date: | 1997-09-12 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility.
Genes Dev., 11, 1997
|
|
6E8D
 
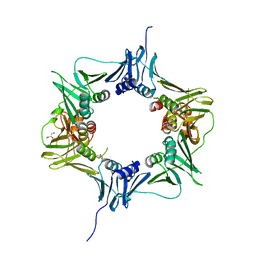 | |
1ZME
 
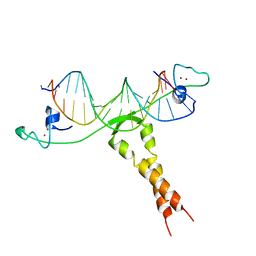 | | CRYSTAL STRUCTURE OF PUT3/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*AP*GP*(5IU)P*TP*GP*GP*CP*TP*(5IU)P*CP*CP*CP*G)-3'), DNA (5'-D(*AP*CP*GP*GP*GP*AP*AP*GP*CP*CP*AP*AP*CP*TP*CP*CP*G)-3'), PROLINE UTILIZATION TRANSCRIPTION ACTIVATOR, ... | | Authors: | Swaminathan, K, Marmorstein, R. | | Deposit date: | 1997-08-06 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a PUT3-DNA complex reveals a novel mechanism for DNA recognition by a protein containing a Zn2Cys6 binuclear cluster.
Nat.Struct.Biol., 4, 1997
|
|
1PXK
 
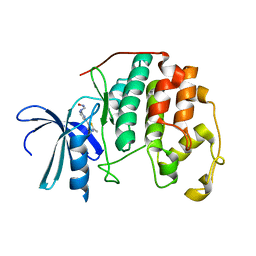 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR N-[4-(2,4-Dimethyl-thiazol-5-yl)pyrimidin-2-yl]-N'-hydroxyiminoformamide | | Descriptor: | Cell division protein kinase 2, N-[4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)PYRIMIDIN-2-YL]-N'-HYDROXYIMIDOFORMAMIDE | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
1XO2
 
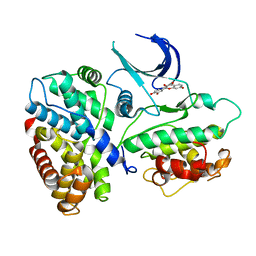 | | Crystal structure of a human cyclin-dependent kinase 6 complex with a flavonol inhibitor, fisetin | | Descriptor: | 3,7,3',4'-TETRAHYDROXYFLAVONE, Cell division protein kinase 6, Cyclin | | Authors: | Lu, H.S, Chang, D.J, Baratte, B, Meijer, L, Schulze-Gahmen, U. | | Deposit date: | 2004-10-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Human Cyclin-Dependent Kinase 6 Complex with a Flavonol Inhibitor, Fisetin.
J.Med.Chem., 48, 2005
|
|
1Q3D
 
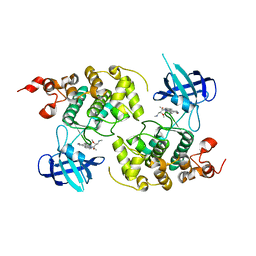 | | GSK-3 Beta complexed with Staurosporine | | Descriptor: | GLYCOGEN SYNTHASE KINASE-3 BETA, STAUROSPORINE | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-07-29 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of the Gsk-3Beta Active Site Using Selective and Non-selective ATP-Mimetic Inhibitors
J.Mol.Biol., 333, 2003
|
|
1PXL
 
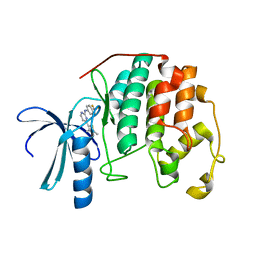 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR [4-(2,4-Dimethyl-thiazol-5-yl)-pyrimidin-2-yl]-(4-trifluoromethyl-phenyl)-amine | | Descriptor: | 4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)-N-[4-(TRIFLUOROMETHYL)PHENYL]PYRIMIDIN-2-AMINE, Cell division protein kinase 2 | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
1Q3W
 
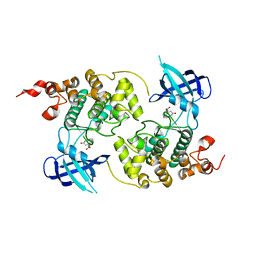 | | GSK-3 Beta complexed with Alsterpaullone | | Descriptor: | 9-NITRO-5,12-DIHYDRO-7H-BENZO[2,3]AZEPINO[4,5-B]INDOL-6-ONE, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-08-01 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of the Gsk-3Beta Active Site Using Selective and Non-selective ATP-Mimetic Inhibitors
J.Mol.Biol., 333, 2003
|
|
1AUZ
 
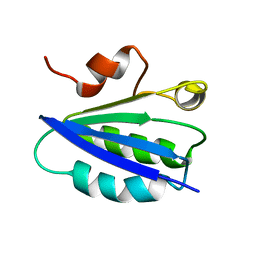 | | SOLUTION STRUCTURE OF SPOIIAA, A PHOSPHORYLATABLE COMPONENT OF THE SYSTEM THAT REGULATES TRANSCRIPTION FACTOR SIGMA-F OF BACILLUS SUBTILIS, NMR, 24 STRUCTURES | | Descriptor: | SPOIIAA | | Authors: | Kovacs, H, Comfort, D, Lord, M, Campbell, I.D, Yudkin, M.D. | | Deposit date: | 1997-09-08 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SpoIIAA, a phosphorylatable component of the system that regulates transcription factor sigmaF of Bacillus subtilis.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1PUE
 
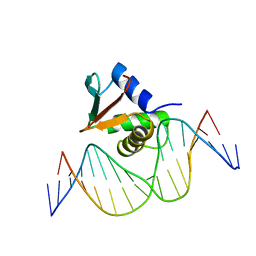 | | PU.1 ETS DOMAIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*GP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*CP*CP*TP*TP*TP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR PU.1 (TF PU.1)) | | Authors: | Kodandapani, R, Pio, F, Ni, C.Z, Piccialli, G, Klemsz, M, McKercher, S, Maki, R.A, Ely, K.R. | | Deposit date: | 1996-07-08 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex.
Nature, 380, 1996
|
|
1ZFS
 
 | |
