1YSN
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL complexed with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-X | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1YX6
 
 | |
1XSC
 
 | | Structure of the nudix enzyme AP4A hydrolase from homo sapiens (E63A mutant) in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bis(5'-nucleosyl)-tetraphosphatase | | Authors: | Swarbrick, J.D, Buyya, S, Gunawardana, D, Gayler, K.R, McLennan, A.G, Gooley, P.R. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and Substrate-binding Mechanism of Human Ap4A Hydrolase
J.Biol.Chem., 280, 2005
|
|
1YE3
 
 | |
1X6M
 
 | | Crystal structure of the glutathione-dependent formaldehyde-activating enzyme (Gfa) | | Descriptor: | GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, SULFATE ION, ... | | Authors: | Neculai, A.M, Neculai, D, Vorholt, J.A, Becker, S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
1XSA
 
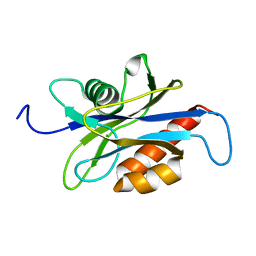 | | Structure of the nudix enzyme AP4A hydrolase from homo sapiens (E63A mutant) | | Descriptor: | Bis(5'-nucleosyl)-tetraphosphatase | | Authors: | Swarbrick, J.D, Buyya, S, Gunawardana, D, Gayler, K.R, McLennan, A.G, Gooley, P.R. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and Substrate-binding Mechanism of Human Ap4A Hydrolase
J.Biol.Chem., 280, 2005
|
|
1XJS
 
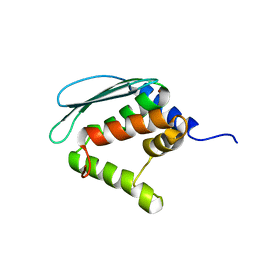 | | Solution structure of Iron-Sulfur cluster assembly protein IscU from Bacillus subtilis, with Zinc bound at the active site. Northeast Structural Genomics Consortium Target SR17 | | Descriptor: | NifU-like protein, ZINC ION | | Authors: | Kornhaber, G.J, Swapna, G.V.T, Ramelot, T.A, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-24 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Iron-Sulfur cluster assembly protein IscU
from Bacillus subtilis, with Zinc bound at the active site.
Northeast Structural Genomics Consortium Target SR17.
To be Published
|
|
1ZXJ
 
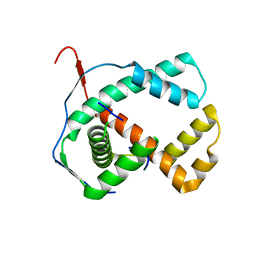 | | Crystal structure of the hypthetical Mycoplasma protein, MPN555 | | Descriptor: | Hypothetical protein MG377 homolog | | Authors: | Schulze-Gahmen, U, Aono, S, Shengfeng, C, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-06-08 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the hypothetical Mycoplasma protein MPN555 suggests a chaperone function.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZTR
 
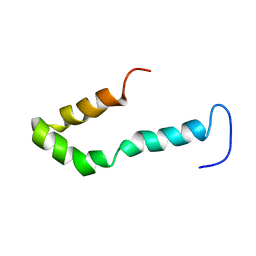 | | Solution structure of Engrailed homeodomain L16A mutant | | Descriptor: | Segmentation polarity homeobox protein engrailed | | Authors: | Religa, T.L, Markson, J.S, Mayor, U, Freund, S.M.V, Fersht, A.R. | | Deposit date: | 2005-05-27 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a protein denatured state and folding intermediate.
Nature, 437, 2005
|
|
1YGO
 
 | |
1YG3
 
 | |
2A55
 
 | | Solution structure of the two N-terminal CCP modules of C4b-binding protein (C4BP) alpha-chain. | | Descriptor: | C4b-binding protein | | Authors: | Jenkins, H.T, Mark, L, Ball, G, Lindahl, G, Uhrin, D, Blom, A.M, Barlow, P.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-12-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Human C4b-binding Protein, Structural Basis for Interaction with Streptococcal M Protein, a Major Bacterial Virulence Factor
J.Biol.Chem., 281, 2006
|
|
2A1C
 
 | | Solution structure of CSP1 | | Descriptor: | CSP1 | | Authors: | Johnsborg, O, Kristiansen, P.E. | | Deposit date: | 2005-06-20 | | Release date: | 2006-05-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Hydrophobic Patch in the Pneumococcal Competence Pheromone CSP is Essential for Specificity and Biological Activity
To be Published
|
|
1YSW
 
 | | Solution structure of the anti-apoptotic protein Bcl-2 complexed with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-2 | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1YSI
 
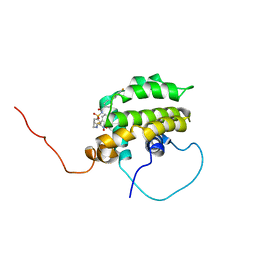 | | Solution structure of the anti-apoptotic protein Bcl-xL in complex with an acyl-sulfonamide-based ligand | | Descriptor: | Apoptosis regulator Bcl-X, N-[(4'-FLUORO-1,1'-BIPHENYL-4-YL)CARBONYL]-3-NITRO-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1YR1
 
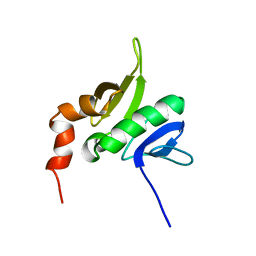 | |
1YSX
 
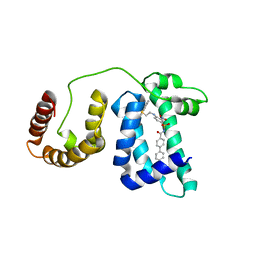 | | Solution structure of domain 3 from human serum albumin complexed to an anti-apoptotic ligand directed against Bcl-xL and Bcl-2 | | Descriptor: | 4-({2-[(2,4-DIMETHYLPHENYL)SULFANYL]ETHYL}AMINO)-N-[(4'-FLUORO-1,1'-BIPHENYL-4-YL)CARBONYL]-3-NITROBENZENESULFONAMIDE, Serum albumin | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
230D
 
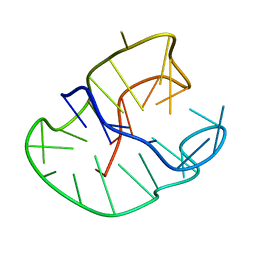 | |
1XSB
 
 | | Structure of the nudix enzyme AP4A hydrolase from homo sapiens (E63A mutant) in complex with ATP. No ATP restraints included | | Descriptor: | Bis(5'-nucleosyl)-tetraphosphatase | | Authors: | Swarbrick, J.D, Buyya, S, Gunawardana, D, Gayler, K.R, McLennan, A.G, Gooley, P.R. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and Substrate-binding Mechanism of Human Ap4A Hydrolase
J.Biol.Chem., 280, 2005
|
|
1Y9H
 
 | | Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 5'-D(*CP*CP*AP*TP*(5CM)P*(BPG)P*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Zhang, N, Lin, C, Huang, X, Kolbanovskiy, A, Hingerty, B.E, Amin, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 2004-12-15 | | Release date: | 2005-03-22 | | Last modified: | 2024-04-24 | | Method: | SOLUTION NMR | | Cite: | Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement.
J.Mol.Biol., 346, 2005
|
|
1YBR
 
 | |
1YWS
 
 | |
1YX4
 
 | |
1YYX
 
 | |
1YWX
 
 | | Solution Structure of Methanococcus maripaludis Protein MMP0443: The Northeast Structural Genomics Consortium Target MrR16 | | Descriptor: | 30S ribosomal protein S24e | | Authors: | Yang, C.S, Acton, T, Shen, Y, Ma, L, Liu, G, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-18 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Methanococcus maripaludis Protein MMP0443: The Northeast Structural Genomics Consortium Target MrR16
To be Published
|
|
