1L36
 
 | |
1L37
 
 | |
1L38
 
 | |
1L39
 
 | |
1L3A
 
 | |
1L3B
 
 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, C2 SPACEGROUP W/ LONG CELL | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-26 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of Mt0146/CbiT Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1L3C
 
 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, C2 SPACEGROUP WITH SHORT CELL | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-26 | | Release date: | 2002-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Crystal Structure of Mt0146/Cbit Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1L3D
 
 | |
1L3E
 
 | | NMR Structures of the HIF-1alpha CTAD/p300 CH1 Complex | | Descriptor: | ZINC ION, hypoxia inducible factor-1 alpha subunit, p300 protein | | Authors: | Freedman, S.J, Sun, Z.J, Poy, F, Kung, A.L, Livingston, D.M, Wagner, G, Eck, M.J. | | Deposit date: | 2002-02-26 | | Release date: | 2002-04-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1L3F
 
 | |
1L3G
 
 | | NMR Structure of the DNA-binding Domain of Cell Cycle Protein, Mbp1(2-124) from Saccharomyces cerevisiae | | Descriptor: | TRANSCRIPTION FACTOR Mbp1 | | Authors: | Nair, M, McIntosh, P.B, Frenkiel, T.A, Kelly, G, Taylor, I.A, Smerdon, S.J, Lane, A.N. | | Deposit date: | 2002-02-27 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the DNA-Binding Domain of the Cell Cycle Protein Mbp1 from Saccharomyces cerevisiae
Biochemistry, 42, 2003
|
|
1L3H
 
 | | NMR structure of P41icf, a potent inhibitor of human cathepsin L | | Descriptor: | MHC CLASS II-ASSOCIATED P41 INVARIANT CHAIN FRAGMENT (P41icf) | | Authors: | Chiva, C, Barthe, P, Codina, A, Giralt, E. | | Deposit date: | 2002-02-27 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Synthesis and NMR structure of P41ICF, a potent inhibitor of human cathepsin L
J.Am.Chem.Soc., 125, 2003
|
|
1L3I
 
 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, ADOHCY BINARY COMPLEX | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-27 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of Mt0146/CbiT Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1L3J
 
 | | Crystal Structure of Oxalate Decarboxylase Formate Complex | | Descriptor: | FORMIC ACID, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Anand, R, Dorrestein, P.C, Kinsland, C, Begley, T.P, Ealick, S.E. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 A resolution.
Biochemistry, 41, 2002
|
|
1L3K
 
 | | UP1, THE TWO RNA-RECOGNITION MOTIF DOMAIN OF HNRNP A1 | | Descriptor: | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN A1 | | Authors: | Vitali, J, Ding, J, Jiang, J, Zhang, Y, Krainer, A.R, Xu, R.-M. | | Deposit date: | 2002-02-27 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Correlated alternative side chain conformations in the RNA-recognition motif of heterogeneous nuclear ribonucleoprotein A1.
Nucleic Acids Res., 30, 2002
|
|
1L3L
 
 | | Crystal structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA | | Descriptor: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, 5'-D(*GP*AP*TP*GP*TP*GP*CP*AP*GP*AP*TP*CP*TP*GP*CP*AP*CP*AP*TP*C)-3', Transcriptional activator protein traR | | Authors: | Zhang, R, Pappas, T, Brace, J.L, Miller, P.C, Oulmassov, T, Molyneaux, J.M, Anderson, J.C, Bashkin, J.K, Winans, S.C, Joachimiak, A. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA.
Nature, 417, 2002
|
|
1L3M
 
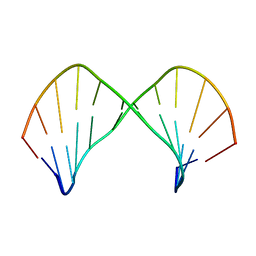 | | The Solution Structure of [d(CGC)r(amamam)d(TTTGCG)]2 | | Descriptor: | 5'-D(*CP*GP*C)-R(P*(A39)P*(A39)P*(A39))-D(P*TP*TP*TP*GP*CP*G)-3' | | Authors: | Tsao, Y.P, Wang, L.Y, Hsu, S.T, Jain, M.L, Chou, S.H, Huang, W.C, Cheng, J.W. | | Deposit date: | 2002-02-28 | | Release date: | 2002-04-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of [d(CGC)r(amamam)d(TTTGCG)]2.
J.Biomol.NMR, 21, 2001
|
|
1L3N
 
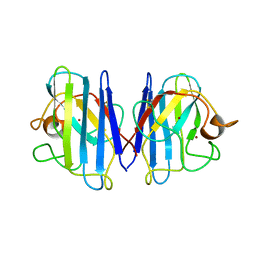 | | The Solution Structure of Reduced Dimeric Copper Zinc SOD: the Structural Effects of Dimerization | | Descriptor: | COPPER (I) ION, ZINC ION, superoxide dismutase [Cu-Zn] | | Authors: | Banci, L, Bertini, I, Cramaro, F, Del Conte, R, Viezzoli, M.S. | | Deposit date: | 2002-02-28 | | Release date: | 2002-05-08 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of reduced dimeric copper zinc superoxide dismutase. The structural effects of dimerization
Eur.J.Biochem., 269, 2002
|
|
1L3O
 
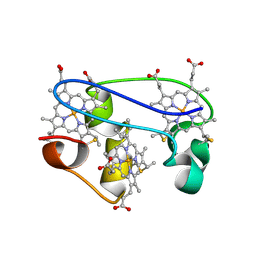 | | SOLUTION STRUCTURE DETERMINATION OF THE FULLY OXIDIZED DOUBLE MUTANT K9-10A CYTOCHROME C7 FROM DESULFUROMONAS ACETOXIDANS, ENSEMBLE OF 35 STRUCTURES | | Descriptor: | HEME C, cytochrome c7 | | Authors: | Assfalg, M, Bertini, I, Turano, P, Bruschi, M, Durand, M.C, Giudici-Orticoni, M.T, Dolla, A. | | Deposit date: | 2002-02-28 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A quick solution structure determination of the fully oxidized double mutant K9-10A cytochrome c7 from Desulfuromonas acetoxidans and mechanistic implications.
J.Biomol.NMR, 22, 2002
|
|
1L3P
 
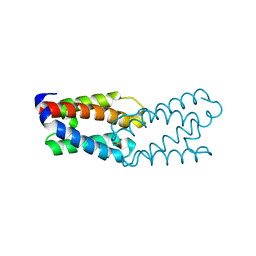 | | CRYSTAL STRUCTURE OF THE FUNCTIONAL DOMAIN OF THE MAJOR GRASS POLLEN ALLERGEN Phl p 5b | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, POLLEN ALLERGEN Phl p 5b | | Authors: | Rajashankar, K.R, Bufe, A, Weber, W, Eschenburg, S, Lindner, B, Betzel, C. | | Deposit date: | 2002-02-28 | | Release date: | 2003-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the functional domain of the major grass-pollen allergen Phlp 5b.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1L3Q
 
 | | H. rufescens abalone shell Lustrin A consensus repeat, FPGKNVNCTSGE, pH 7.4, 1-H NMR structure | | Descriptor: | Lustrin A | | Authors: | Evans, J.S, Wustman, B.A, Zhang, B, Morse, D.E. | | Deposit date: | 2002-02-28 | | Release date: | 2002-03-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Model peptide studies of sequence regions in the elastomeric biomineralization protein, Lustrin A. I. The C-domain consensus-PG-, -NVNCT-motif
Biopolymers, 63, 2002
|
|
1L3R
 
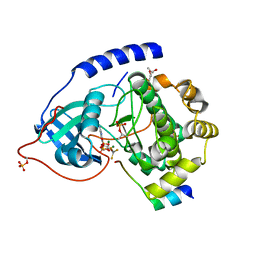 | | Crystal Structure of a Transition State Mimic of the Catalytic Subunit of cAMP-dependent Protein Kinase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Madhusudan, Akamine, P, Xuong, N.-H, Taylor, S.S. | | Deposit date: | 2002-02-28 | | Release date: | 2002-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase.
Nat.Struct.Biol., 9, 2002
|
|
1L3S
 
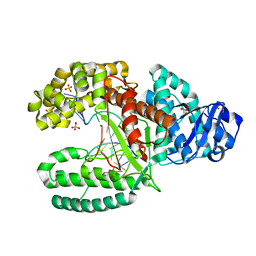 | | Crystal Structure of Bacillus DNA Polymerase I Fragment complexed to 9 base pairs of duplex DNA. | | Descriptor: | 5'-D(*GP*A*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*G)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-01 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1L3T
 
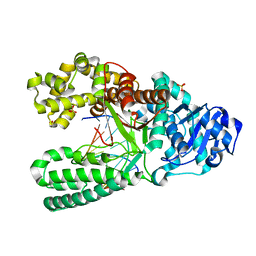 | | Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 10 base pairs of duplex DNA following addition of a single dTTP residue | | Descriptor: | 5'-D(*GP*AP*CP*G*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-01 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1L3U
 
 | | Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 11 base pairs of duplex DNA following addition of a dTTP and a dATP residue. | | Descriptor: | 5'-D(*GP*AP*C*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*A)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-01 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
