7JN1
 
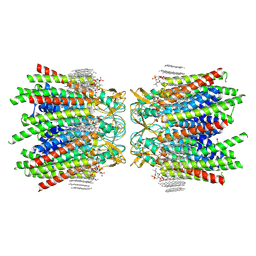 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-08-03 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
8A3D
 
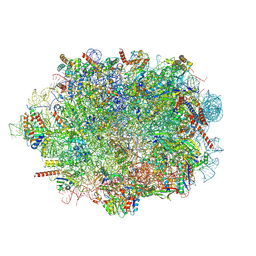 | | Human mature large subunit of the ribosome with eIF6 and homoharringtonine bound | | Descriptor: | (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 28S ribosomal RNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Faille, A, Warren, A.J, Dent, K.C. | | Deposit date: | 2022-06-08 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | The chemical landscape of the human ribosome at 1.67 angstrom resolution.
Biorxiv, 2023
|
|
1GU4
 
 | |
1GU5
 
 | |
1GTW
 
 | |
3H7D
 
 | | The crystal structure of the cathepsin K Variant M5 in complex with chondroitin-4-sulfate | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, CALCIUM ION, Cathepsin K, ... | | Authors: | Cherney, M.M, Kienetz, M, Bromme, D, James, M.N.G. | | Deposit date: | 2009-04-24 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structure-activity analysis of cathepsin K/chondroitin 4-sulfate interactions.
J.Biol.Chem., 286, 2011
|
|
8TEK
 
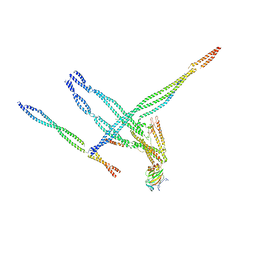 | | Baseplate of Nexin-dynein regulatory complex from Tetrahymena thermophila | | Descriptor: | CFAP20, Cilia- and flagella-associated protein 91, Coiled-coil protein, ... | | Authors: | Ghanaeian, A.G, Black, C.S, Yang, S.K, Bui, K.H. | | Deposit date: | 2023-07-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Integrated modeling of the Nexin-dynein regulatory complex reveals its regulatory mechanism.
Nat Commun, 14, 2023
|
|
7DBP
 
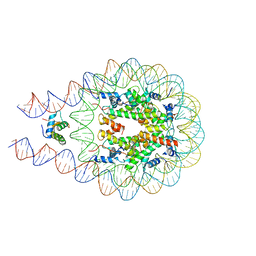 | | Linker histone defines structure and self-association behaviour of the 177 bp human chromosome | | Descriptor: | DNA (175-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | Authors: | Wang, S, Vogirala, K.V, Soman, A, Liu, Z.B. | | Deposit date: | 2020-10-21 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Linker histone defines structure and self-association behaviour of the 177 bp human chromatosome.
Sci Rep, 11, 2021
|
|
8GBK
 
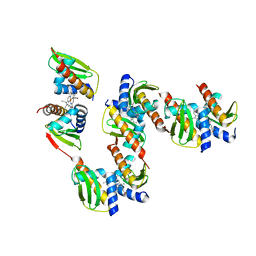 | |
7OVS
 
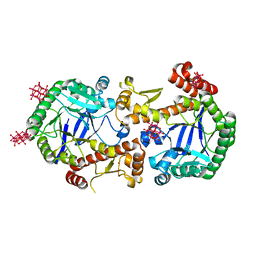 | | Heterodimeric murine tRNA-guanine transglycosylase in the presence of Anderson-Evans type (TEW) and Strandberg type polyoxometalate (POM) | | Descriptor: | 6-tungstotellurate(VI), Queuine tRNA-ribosyltransferase accessory subunit 2, Queuine tRNA-ribosyltransferase catalytic subunit 1, ... | | Authors: | Sebastiani, M, Heine, A, Reuter, K. | | Deposit date: | 2021-06-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Acs Chem.Biol., 17, 2022
|
|
4V7C
 
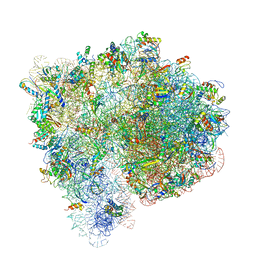 | | Structure of the Ribosome with Elongation Factor G Trapped in the Pre-Translocation State (pre-translocation 70S*tRNA structure) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Brilot, A.F, Korostelev, A.A, Ermolenko, D.N, Grigorieff, N. | | Deposit date: | 2013-11-20 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the ribosome with elongation factor G trapped in the pretranslocation state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7OJU
 
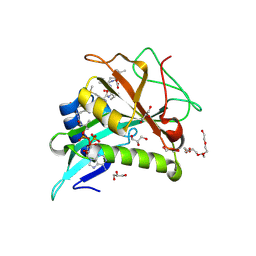 | | Chaetomium thermophilum Naa50 GNAT-domain in complex with bisubstrate analogue CoA-Ac-MVNAL | | Descriptor: | CARBOXYMETHYL COENZYME *A, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Weidenhausen, J, Kopp, J, Sinning, I. | | Deposit date: | 2021-05-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Extended N-Terminal Acetyltransferase Naa50 in Filamentous Fungi Adds to Naa50 Diversity.
Int J Mol Sci, 23, 2022
|
|
8GXX
 
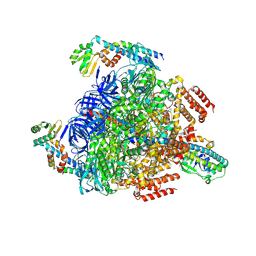 | | 3 nucleotide-bound V1EG of V/A-ATPase from Thermus thermophilus. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8GXU
 
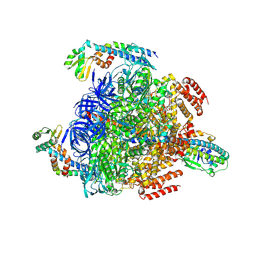 | | 1 ATP-bound V1EG of V/A-ATPase from Thermus thermophilus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, SULFATE ION, V-type ATP synthase alpha chain, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8GXY
 
 | | 2 sulfate-bound V1EG of V/A-ATPase from Thermus thermophilus. | | Descriptor: | SULFATE ION, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8GXZ
 
 | | 1 sulfate and 1 ATP bound V1EG of V/A-ATPase from Thermus thermophilus. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8GXW
 
 | | 2 ATP-bound V1EG of V/A-ATPase from Thermus thermophilus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8H3X
 
 | | Bacteroide Fragilis Toxin in complex with nanobody 282 | | Descriptor: | Fragilysin, ZINC ION, nanobody 282 | | Authors: | Wen, Y, Guo, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Screening and epitope characterization of diagnostic nanobody against total and activated Bacteroides fragilis toxin.
Front Immunol, 14, 2023
|
|
7ONM
 
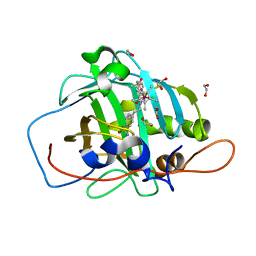 | | Carbonic anhydrase II mutant (N67G-E69R-I91C) dually binding an IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(4-azanyl-9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
7ONQ
 
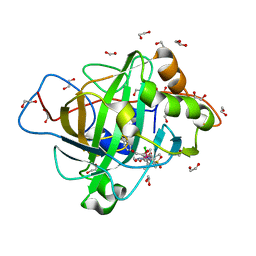 | | Carbonic anhydrase II mutant (E69C) dually binding an IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(4-azanyl-9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
7ONV
 
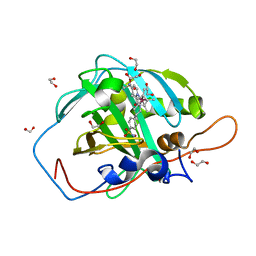 | | Carbonic anhydrase II mutant (I91C) dually binding an IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(4-azanyl-9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-26 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
6XYW
 
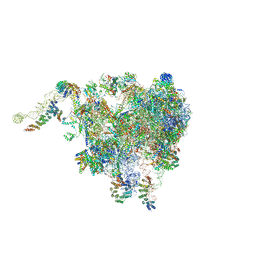 | | Structure of the plant mitochondrial ribosome | | Descriptor: | 28S ribosomal S34 protein, 3-hydroxyisobutyryl-CoA hydrolase-like protein 2, mitochondrial, ... | | Authors: | Soufari, H, Waltz, F, Bochler, A, Giege, P, Hashem, Y. | | Deposit date: | 2020-01-31 | | Release date: | 2020-04-15 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM structure of the RNA-rich plant mitochondrial ribosome.
Nat.Plants, 6, 2020
|
|
4UUZ
 
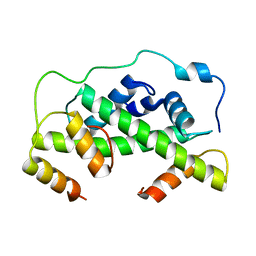 | | MCM2-histone complex | | Descriptor: | DNA REPLICATION LICENSING FACTOR MCM2, HISTONE H3, HISTONE H4 | | Authors: | Richet, N, Liu, D, Legrand, P, Bakail, M, Compper, C, Besle, A, Guerois, R, Ochsenbein, F. | | Deposit date: | 2014-08-01 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insight Into How the Human Helicase Subunit Mcm2 May Act as a Histone Chaperone Together with Asf1 at the Replication Fork.
Nucleic Acids Res., 43, 2015
|
|
8HME
 
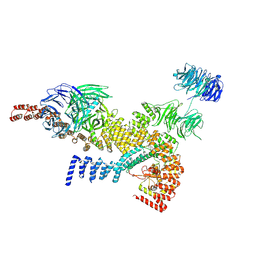 | | head module state 1 of Tetrahymena IFT-A | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transporter, WD40 repeat protein, ... | | Authors: | Ma, Y, Wu, J, Lei, M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insight into the intraflagellar transport complex IFT-A and its assembly in the anterograde IFT train.
Nat Commun, 14, 2023
|
|
4V7D
 
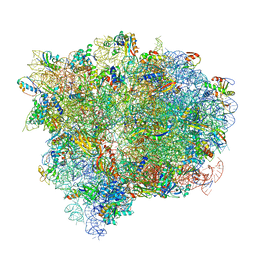 | | Structure of the Ribosome with Elongation Factor G Trapped in the Pre-Translocation State (pre-translocation 70S*tRNA*EF-G structure) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Brilot, A.F, Korostelev, A.A, Ermolenko, D.N, Grigorieff, N. | | Deposit date: | 2013-11-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the ribosome with elongation factor G trapped in the pretranslocation state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
