3BXF
 
 | | Crystal structure of effector binding domain of central glycolytic gene regulator (CggR) from Bacillus subtilis in complex with effector fructose-1,6-bisphosphate | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, 1,6-di-O-phosphono-beta-D-fructofuranose, CHLORIDE ION, ... | | Authors: | Rezacova, P, Otwinowski, Z. | | Deposit date: | 2008-01-13 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the effector-binding domain of repressor Central glycolytic gene Regulator from Bacillus subtilis reveal ligand-induced structural changes upon binding of several glycolytic intermediates.
Mol.Microbiol., 69, 2008
|
|
3BM1
 
 | |
3G5W
 
 | | Crystal structure of Blue Copper Oxidase from Nitrosomonas europaea | | Descriptor: | COPPER (II) ION, CU-O LINKAGE, CU-O-CU LINKAGE, ... | | Authors: | Lawton, T.J, Sayavedra-Soto, L.A, Arp, D.J, Rosenzweig, A.C. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a two-domain multicopper oxidase: implications for the evolution of multicopper blue proteins.
J.Biol.Chem., 284, 2009
|
|
5DHV
 
 | | HIV-1 Rev NTD dimers with variable crossing angles | | Descriptor: | Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, light chain, ... | | Authors: | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | Deposit date: | 2015-08-31 | | Release date: | 2016-06-22 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
3TW2
 
 | | High resolution structure of human histidine triad nucleotide-binding protein 1 (hHINT1)/AMP complex in a monoclinic space group | | Descriptor: | ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Dolot, R.M, Wlodarczyk, A, Ozga, M, Krakowiak, A, Nawrot, B. | | Deposit date: | 2011-09-21 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A new crystal form of human histidine triad nucleotide-binding protein 1 (hHINT1) in complex with adenosine 5'-monophosphate at 1.38 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4ZEV
 
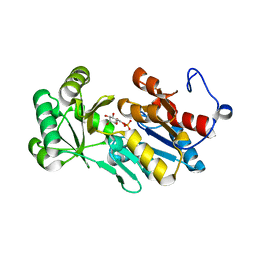 | | Crystal structure of PfHAD1 in complex with mannose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-mannopyranose, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2015-04-20 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-domain closure enables diverse substrate recognition by the C2-type haloacid dehalogenase-like sugar phosphatase Plasmodium falciparum HAD1.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
1H0J
 
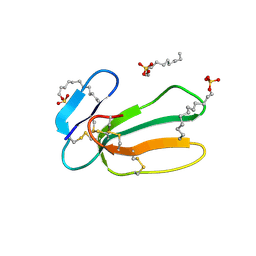 | | Structural Basis of the Membrane-induced Cardiotoxin A3 Oligomerization | | Descriptor: | CARDIOTOXIN-3, DODECYL SULFATE | | Authors: | Forouhar, F, Huang, W.-N, Liu, J.-H, Chien, K.-Y, Wu, W.-G, Hsiao, C.-D. | | Deposit date: | 2002-06-20 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Membrane-Induced Cardiotoxin A3 Oligomerization
J.Biol.Chem., 278, 2003
|
|
3BIS
 
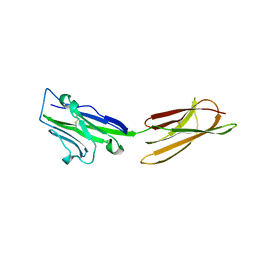 | | Crystal Structure of the PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3RC7
 
 | |
2BIS
 
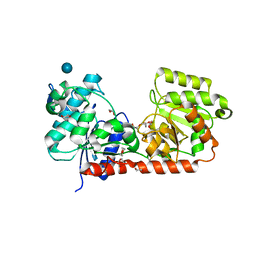 | | Structure of glycogen synthase from Pyrococcus abyssi | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLGA GLYCOGEN SYNTHASE, GLYCEROL, ... | | Authors: | Horcajada, C, Guinovart, J.J, Fita, I, Ferrer, J.C. | | Deposit date: | 2005-01-25 | | Release date: | 2005-11-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of an Archaeal Glycogen Synthase: Insights Into Oligomerisation and Substrate Binding of Eukaryotic Glycogen Synthases.
J.Biol.Chem., 281, 2006
|
|
5DCH
 
 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I in complex with MIPS-0000851 (3-[(2-METHYLBENZYL)SULFANYL]-4H-1,2,4-TRIAZOL-4-AMINE) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine, GLYCEROL, ... | | Authors: | McMahon, R.M, Martin, J.L. | | Deposit date: | 2015-08-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
1HFH
 
 | | SOLUTION STRUCTURE OF A PAIR OF COMPLEMENT MODULES BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | FACTOR H, 15TH AND 16TH C-MODULE PAIR | | Authors: | Barlow, P.N, Steinkasserer, A, Norman, D.G, Kieffer, B, Wiles, A.P, Sim, R.B, Campbell, I.D. | | Deposit date: | 1993-02-23 | | Release date: | 1993-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of complement modules by nuclear magnetic resonance.
J.Mol.Biol., 232, 1993
|
|
1HFI
 
 | | SOLUTION STRUCTURE OF A PAIR OF COMPLEMENT MODULES BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | FACTOR H, 15TH C-MODULE PAIR | | Authors: | Barlow, P.N, Steinkasserer, A, Norman, D.G, Kieffer, B, Wiles, A.P, Sim, R.B, Campbell, I.D. | | Deposit date: | 1993-02-23 | | Release date: | 1993-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of complement modules by nuclear magnetic resonance.
J.Mol.Biol., 232, 1993
|
|
1GWY
 
 | | Crystal structure of the water-soluble state of the pore-forming cytolysin Sticholysin II | | Descriptor: | STICHOLYSIN II, SULFATE ION | | Authors: | Mancheno, J.M, Martin-Benito, J, Martinez-Ripoll, M, Gavilanes, J.G, Hermoso, J.A. | | Deposit date: | 2002-03-26 | | Release date: | 2003-06-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal and Electron Microscopy Structures of Sticholysin II Actinoporin Reveal Insights Into the Mechanism of Membrane Pore Formation
Structure, 11, 2003
|
|
2OR3
 
 | |
1GUB
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | Descriptor: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, NICKEL (II) ION | | Authors: | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2002-01-24 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
3BXG
 
 | |
5M3Q
 
 | |
3BKB
 
 | | Crystal structure of human Feline Sarcoma Viral Oncogene Homologue (v-FES) | | Descriptor: | 1,2-ETHANEDIOL, Proto-oncogene tyrosine-protein kinase Fes/Fps, STAUROSPORINE, ... | | Authors: | Filippakopoulos, P, Salah, E, Fedorov, O, Cooper, C, Ugochukwu, E, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Coupling of SH2-Kinase Domains Links Fes and Abl Substrate Recognition and Kinase Activation
Cell(Cambridge,Mass.), 134, 2008
|
|
1GUD
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | Descriptor: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, ZINC ION | | Authors: | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2002-01-24 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
1GYK
 
 | | Serum Amyloid P Component co-crystallised with MOBDG at neutral pH | | Descriptor: | CALCIUM ION, METHYL 4,6-O-[(1R)-1-CARBOXYETHYLIDENE]-BETA-D-GALACTOPYRANOSIDE, SERUM AMYLOID P-COMPONENT | | Authors: | Thompson, D, Pepys, M.B, Tickle, I, Wood, S.P. | | Deposit date: | 2002-04-25 | | Release date: | 2003-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structures of Crystalline Complexes of Human Serum Amyloid P Component with its Carbohydrate Ligand, the Cyclic Pyruvate Acetal of Galactose
J.Mol.Biol., 320, 2002
|
|
5AYY
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE REACTANT QUINOLINATE | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating], QUINOLINIC ACID | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
2BB6
 
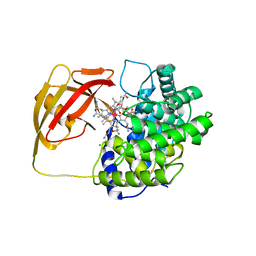 | | Structure of Cobalamin-complexed Bovine Transcobalamin in Monoclinic Crystal Form | | Descriptor: | CHLORIDE ION, COBALAMIN, Transcobalamin II | | Authors: | Wuerges, J, Garau, G, Geremia, S, Fedosov, S.N, Petersen, T.E, Randaccio, L. | | Deposit date: | 2005-10-17 | | Release date: | 2006-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for mammalian vitamin B12 transport by transcobalamin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5B26
 
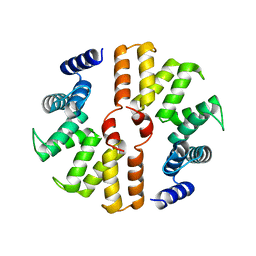 | | Crystal structure of mouse SEL1L | | Descriptor: | Protein sel-1 homolog 1 | | Authors: | Jeong, H, Lee, C. | | Deposit date: | 2016-01-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of SEL1L: Insight into the roles of SLR motifs in ERAD pathway
Sci Rep, 6, 2016
|
|
5Z67
 
 | |
