8B2L
 
 | | Cryo-EM structure of the plant 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 30S ribosomal protein S15, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-09-14 | | Release date: | 2023-08-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
7OHW
 
 | |
7OHP
 
 | |
7OHV
 
 | |
7OHX
 
 | |
7OHR
 
 | | Nog1-TAP associated immature ribosomal particle population E from S. cerevisiae | | Descriptor: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Milkereit, P, Poell, G. | | Deposit date: | 2021-05-11 | | Release date: | 2021-11-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.72 Å) | | Cite: | Analysis of subunit folding contribution of three yeast large ribosomal subunit proteins required for stabilisation and processing of intermediate nuclear rRNA precursors.
Plos One, 16, 2021
|
|
5MIO
 
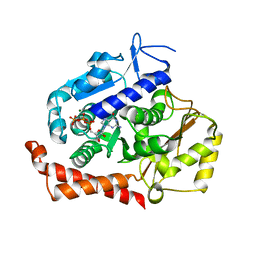 | | KIF2C-DARPIN FUSION PROTEIN BOUND TO TUBULIN | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF2C,KIF2C FUSED TO A DARPIN,KIF2C FUSED TO A DARPIN, ... | | Authors: | Wang, W, Gigant, B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Insight into microtubule disassembly by kinesin-13s from the structure of Kif2C bound to tubulin.
Nat Commun, 8, 2017
|
|
7O80
 
 | | Rabbit 80S ribosome in complex with eRF1 and ABCE1 stalled at the STOP codon in the mutated SARS-CoV-2 slippery site | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
4KZY
 
 | |
6TH4
 
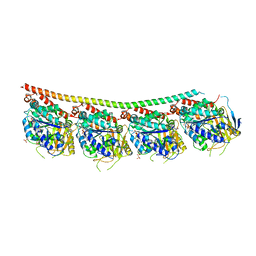 | | Tubulin-inhibitor complex | | Descriptor: | 1,2,3,9-tetramethoxy-6-methylidene-5~{H}-cyclohepta[a]naphthalen-8-one, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Varela, P.F, Gigant, B. | | Deposit date: | 2019-11-18 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | B-nor-methylene Colchicinoid PT-100 Selectively Induces Apoptosis in Multidrug-Resistant Human Cancer Cells via an Intrinsic Pathway in a Caspase-Independent Manner
Acs Omega, 7, 2022
|
|
6FQ6
 
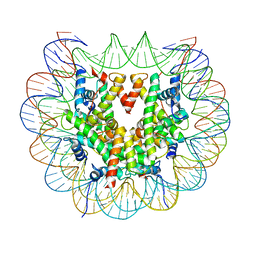 | | Class 2 : distorted nucleosome | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B, ... | | Authors: | Bilokapic, S, Halic, M. | | Deposit date: | 2018-02-13 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural rearrangements of the histone octamer translocate DNA.
Nat Commun, 9, 2018
|
|
3GQP
 
 | |
3GYS
 
 | |
3GQR
 
 | |
1R00
 
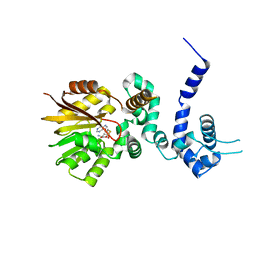 | | Crystal structure of aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-homocysteine (SAH) | | Descriptor: | ACETATE ION, S-ADENOSYL-L-HOMOCYSTEINE, aclacinomycin-10-hydroxylase | | Authors: | Jansson, A, Niemi, J, Lindqvist, Y, Mantsala, P, Schneider, G. | | Deposit date: | 2003-09-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Aclacinomycin-10-Hydroxylase, a S-Adenosyl-L-Methionine-dependent Methyltransferase Homolog Involved in Anthracycline Biosynthesis in Streptomyces purpurascens.
J.Mol.Biol., 334, 2003
|
|
1R05
 
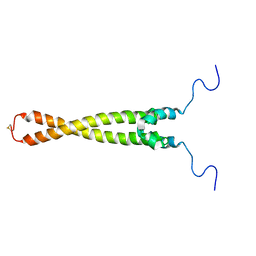 | | Solution Structure of Max B-HLH-LZ | | Descriptor: | Max protein | | Authors: | Sauv, S, Tremblay, L, Lavigne, P. | | Deposit date: | 2003-09-19 | | Release date: | 2003-10-21 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of a mutant of the Max b/HLH/LZ free of DNA: insights into the specific and reversible DNA binding mechanism of dimeric transcription factors
J.Mol.Biol., 342, 2004
|
|
1QND
 
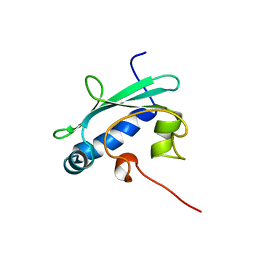 | | STEROL CARRIER PROTEIN-2, NMR, 20 STRUCTURES | | Descriptor: | NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Lopez-Garcia, F, Szyperski, T, Dyer, J.H, Choinowski, T, Seedorf, U, Hauser, H, Wuthrich, K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-07-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Sterol Carrier Protein-2: Implications for the Biological Role
J.Mol.Biol., 295, 2000
|
|
1QBE
 
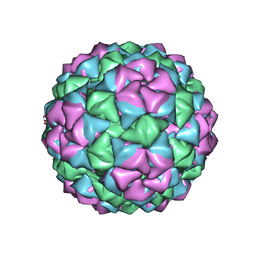 | | BACTERIOPHAGE Q BETA CAPSID | | Descriptor: | BACTERIOPHAGE Q BETA CAPSID | | Authors: | Liljas, L, Golmohammadi, R. | | Deposit date: | 1996-01-10 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The crystal structure of bacteriophage Q beta at 3.5 A resolution.
Structure, 4, 1996
|
|
1R09
 
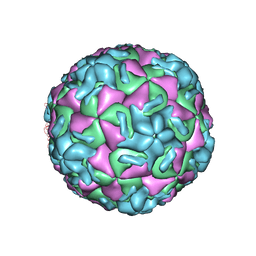 | | HUMAN RHINOVIRUS 14 COMPLEXED WITH ANTIVIRAL COMPOUND R 61837 | | Descriptor: | 3-METHOXY-6-[4-(3-METHYLPHENYL)-1-PIPERAZINYL]PYRIDAZINE, DIMETHYL SULFOXIDE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), ... | | Authors: | Chapman, M.S, Minor, I, Rossmann, M.G, Diana, G.D, Andries, K. | | Deposit date: | 1990-05-04 | | Release date: | 1991-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Human rhinovirus 14 complexed with antiviral compound R 61837.
J.Mol.Biol., 217, 1991
|
|
1QMN
 
 | |
1QMV
 
 | | thioredoxin peroxidase B from red blood cells | | Descriptor: | PEROXIREDOXIN-2 | | Authors: | Isupov, M.N, Littlechild, J.A, Lebedev, A.A, Errington, N, Vagin, A.A, Schroder, E. | | Deposit date: | 1999-10-07 | | Release date: | 2000-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Decameric 2-Cys Peroxiredoxin from Human Erythrocytes at 1.7 A Resolution.
Structure, 8, 2000
|
|
1QNL
 
 | |
1QO4
 
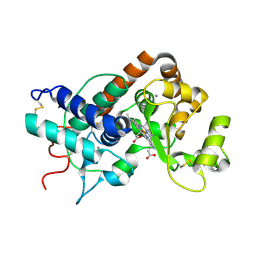 | | ARABIDOPSIS THALIANA PEROXIDASE A2 AT ROOM TEMPERATURE | | Descriptor: | CALCIUM ION, PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Henriksen, A, Gajhede, M. | | Deposit date: | 1999-11-02 | | Release date: | 2000-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Differential Activity and Structure of Highly Similar Peroxidases. Spectroscopic, Crystallographic, and Enzymatic Analyses of Lignifying Arabidopsis Thaliana Peroxidase A2 and Horseradish Peroxidase A2
Biochemistry, 40, 2001
|
|
1R2S
 
 | |
3LUJ
 
 | |
