3ASR
 
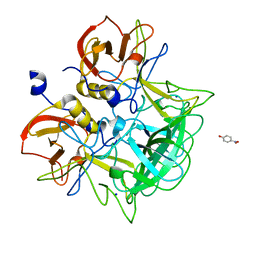 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with Lewis-a | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3BE8
 
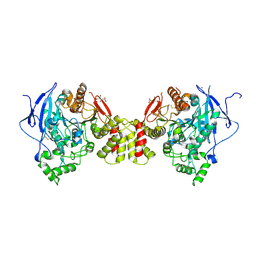 | | Crystal structure of the synaptic protein neuroligin 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Fabrichny, I.P, Leone, P, Sulzenbacher, G, Comoletti, D, Miller, M.T, Taylor, P, Bourne, Y, Marchot, P. | | Deposit date: | 2007-11-16 | | Release date: | 2008-01-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of the Synaptic Protein Neuroligin and Its beta-Neurexin Complex: Determinants for Folding and Cell Adhesion
Neuron, 56, 2007
|
|
3CNI
 
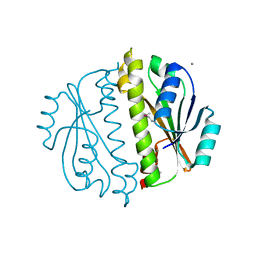 | | Crystal structure of a domain of a putative ABC type-2 transporter from Thermotoga maritima MSB8 | | Descriptor: | CALCIUM ION, Putative ABC type-2 transporter | | Authors: | Filippova, E.V, Shumilin, I, Tkaczuk, K.L, Cymborowski, M, Chruszcz, M, Xu, X, Que, Q, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of the putative ABC-type 2 transporter from Thermotoga maritima MSB8.
J.Struct.Funct.Genom., 15, 2014
|
|
1AK9
 
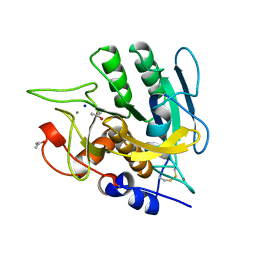 | | SUBTILISIN MUTANT 8321 | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SODIUM ION, ... | | Authors: | Whitlow, M, Howard, A.J, Wood, J.F. | | Deposit date: | 1997-05-30 | | Release date: | 1997-11-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stability for subtilisin BPN' through incremental changes in the free energy of unfolding.
Biochemistry, 28, 1989
|
|
1C21
 
 | | E. COLI METHIONINE AMINOPEPTIDASE: METHIONINE COMPLEX | | Descriptor: | COBALT (II) ION, METHIONINE, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Lowther, W.T, Zhang, Y, Sampson, P.B, Honek, J.F, Matthews, B.W. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of Escherichia coli methionine aminopeptidase from the structural analysis of reaction products and phosphorus-based transition-state analogues.
Biochemistry, 38, 1999
|
|
1C4V
 
 | | SELECTIVE NON ELECTROPHILIC THROMBIN INHIBITORS WITH CYCLOHEXYL MOIETIES. | | Descriptor: | 2-(2,2-DIPHENYL-ETHYL)-7-METHYL-1,3-DIOXO-2,3,5,8-TETRAHYDRO-1H-[1,2,4]TRIAZOLO [1,2-A]PYRIDAZINE-5-CARBOXYLIC ACID(4-CARBAMIMIDOYL-CYCLOHEXYLMETHYL)-AMIDE, HIRUGEN, SODIUM ION, ... | | Authors: | Krishnan, R, Mochalkin, I, Arni, R.K, Tulinsky, A. | | Deposit date: | 1999-09-25 | | Release date: | 2000-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of thrombin complexed with selective non-electrophilic inhibitors having cyclohexyl moieties at P1.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1C4U
 
 | | SELECTIVE NON ELECTROPHILIC THROMBIN INHIBITORS WITH CYCLOHEXYL MOIETIES. | | Descriptor: | 2-[2-(4-BROMO-BENZENESULFONYL)-ETHYL]-1-3-DIOXO-2,3,5,8-TETRAHYDRO-1H-[1,2,4]TRIAZOLO[1,2-A]PYRIDAZINE-5-CARBOXYLIC ACID(4-CARBAMIMIDOYL-CYCLOHEXYLMETHYL)-AMIDE, PROTEIN (HIRUGEN), SODIUM ION, ... | | Authors: | Krishnan, R, Mochalkin, I, Arni, R.K, Tulinsky, A. | | Deposit date: | 1999-09-25 | | Release date: | 2000-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of thrombin complexed with selective non-electrophilic inhibitors having cyclohexyl moieties at P1.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1C5L
 
 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | CALCIUM ION, Hirudin, SODIUM ION, ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5N
 
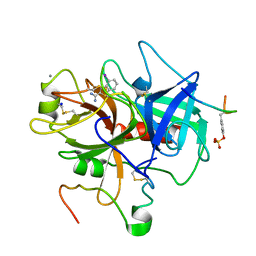 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | 4-IODOBENZO[B]THIOPHENE-2-CARBOXAMIDINE, CALCIUM ION, Hirudin, ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5O
 
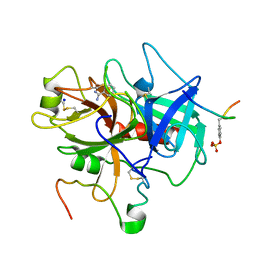 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | BENZAMIDINE, Hirudin, SODIUM ION, ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C8V
 
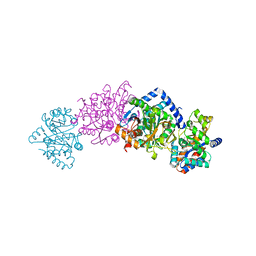 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-HYDROXYPHENYLTHIO)-BUTYLPHOSPHONIC ACID | | Descriptor: | 4-(2-HYDROXYPHENYLTHIO)-1-BUTENYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-07-30 | | Release date: | 2000-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
1C10
 
 | | CRYSTAL STRUCTURE OF HEW LYSOZYME UNDER PRESSURE OF XENON (8 BAR) | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S, Bourguet, W, Fourme, R. | | Deposit date: | 1999-07-16 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
1C24
 
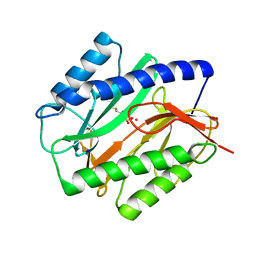 | | E. COLI METHIONINE AMINOPEPTIDASE: METHIONINE PHOSPHINATE COMPLEX | | Descriptor: | (1-AMINO-3-METHYLSULFANYL-PROPYL)-PHOSPHINIC ACID, COBALT (II) ION, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Lowther, W.T, Zhang, Y, Sampson, P.B, Honek, J.F, Matthews, B.W. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the mechanism of Escherichia coli methionine aminopeptidase from the structural analysis of reaction products and phosphorus-based transition-state analogues.
Biochemistry, 38, 1999
|
|
1BPH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | Descriptor: | 1,2-DICHLOROETHANE, INSULIN A CHAIN (PH 9), INSULIN B CHAIN (PH 9), ... | | Authors: | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | Deposit date: | 1992-10-30 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
1CK7
 
 | | GELATINASE A (FULL-LENGTH) | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROTEIN (GELATINASE A), ... | | Authors: | Morgunova, E, Tuuttila, A, Bergmann, U, Isupov, M, Lindqvist, Y, Schneider, G, Tryggvason, K. | | Deposit date: | 1999-04-28 | | Release date: | 1999-08-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human pro-matrix metalloproteinase-2: activation mechanism revealed.
Science, 284, 1999
|
|
1CQX
 
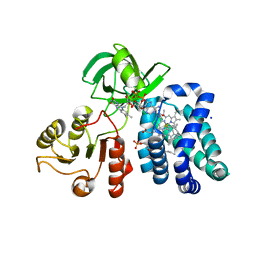 | | Crystal structure of the flavohemoglobin from Alcaligenes eutrophus at 1.75 A resolution | | Descriptor: | 1-[GLYCEROLYLPHOSPHONYL]-2-[8-(2-HEXYL-CYCLOPROPYL)-OCTANAL-1-YL]-3-[HEXADECANAL-1-YL]-GLYCEROL, FLAVIN-ADENINE DINUCLEOTIDE, FLAVOHEMOPROTEIN, ... | | Authors: | Ermler, U, Siddiqui, R.A, Cramm, R, Friedrich, B. | | Deposit date: | 1999-08-12 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the flavohemoglobin from Alcaligenes eutrophus at 1.75 A resolution.
EMBO J., 14, 1995
|
|
1D7V
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH NMA | | Descriptor: | N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-2-METHYLALANINE, POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
1A2C
 
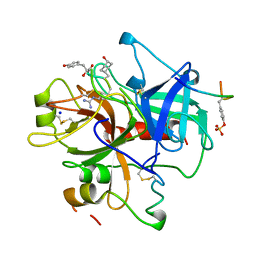 | | Structure of thrombin inhibited by AERUGINOSIN298-A from a BLUE-GREEN ALGA | | Descriptor: | Aeruginosin 298-A, Hirudin variant-2, SODIUM ION, ... | | Authors: | Rios-Steiner, J.L, Murakami, M, Tulinsky, A. | | Deposit date: | 1997-12-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Thrombin Inhibited by Aeruginosin 298-A from a Blue-Green Alga
J.Am.Chem.Soc., 120, 1998
|
|
1D7R
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH 5PA | | Descriptor: | N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID, POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
1D7U
 
 | | Crystal structure of the complex of 2,2-dialkylglycine decarboxylase with LCS | | Descriptor: | POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), SODIUM ION, ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
1DI5
 
 | |
1C23
 
 | | E. COLI METHIONINE AMINOPEPTIDASE: METHIONINE PHOSPHONATE COMPLEX | | Descriptor: | (1-AMINO-3-METHYLSULFANYL-PROPYL)-PHOSPHONIC ACID, COBALT (II) ION, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Lowther, W.T, Zhang, Y, Sampson, P.B, Honek, J.F, Matthews, B.W. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the mechanism of Escherichia coli methionine aminopeptidase from the structural analysis of reaction products and phosphorus-based transition-state analogues.
Biochemistry, 38, 1999
|
|
1C22
 
 | | E. COLI METHIONINE AMINOPEPTIDASE: TRIFLUOROMETHIONINE COMPLEX | | Descriptor: | 2-AMINO-4-TRIFLUOROMETHYLSULFANYL-BUTYRIC ACID, COBALT (II) ION, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Lowther, W.T, Zhang, Y, Sampson, P.B, Honek, J.F, Matthews, B.W. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into the mechanism of Escherichia coli methionine aminopeptidase from the structural analysis of reaction products and phosphorus-based transition-state analogues.
Biochemistry, 38, 1999
|
|
1C27
 
 | | E. COLI METHIONINE AMINOPEPTIDASE:NORLEUCINE PHOSPHONATE COMPLEX | | Descriptor: | (1-AMINO-PENTYL)-PHOSPHONIC ACID, COBALT (II) ION, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Lowther, W.T, Zhang, Y, Sampson, P.B, Honek, J.F, Matthews, B.W. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the mechanism of Escherichia coli methionine aminopeptidase from the structural analysis of reaction products and phosphorus-based transition-state analogues.
Biochemistry, 38, 1999
|
|
1DPH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | Descriptor: | 1,2-DICHLOROETHANE, INSULIN A CHAIN (PH 11), INSULIN B CHAIN (PH 11), ... | | Authors: | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | Deposit date: | 1992-10-30 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
