4EAG
 
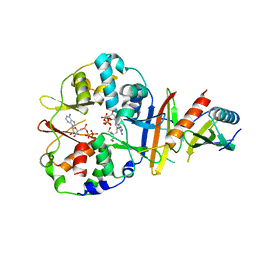 | | Co-crystal structure of an chimeric AMPK core with ATP | | Descriptor: | 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
4EB0
 
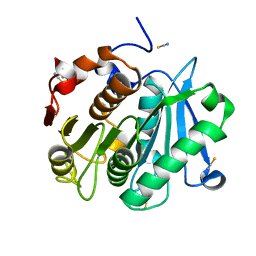 | | Crystal structure of Leaf-branch compost bacterial cutinase homolog | | Descriptor: | LCC, THIOCYANATE ION | | Authors: | Sulaiman, S, You, D.J, Eiko, K, Koga, Y, Kanaya, S. | | Deposit date: | 2012-03-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Leaf-branch compost bacterial cutinase homolog
To be Published
|
|
4EBU
 
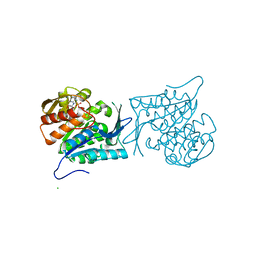 | | Crystal structure of a sugar kinase (Target EFI-502312) from Oceanicola granulosus, with bound AMP/ADP crystal form I | | Descriptor: | 2-dehydro-3-deoxygluconokinase, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-24 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a sugar kinase (Target EFI-502312) from Oceanicola granulosus, with bound AMP/ADP crystal form I
TO BE PUBLISHED
|
|
4E43
 
 | |
4EDG
 
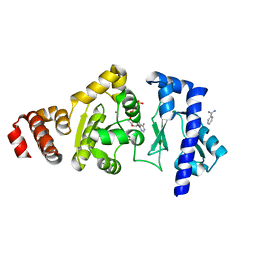 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to ATP and Manganese | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BENZAMIDINE, DNA primase, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
4EFC
 
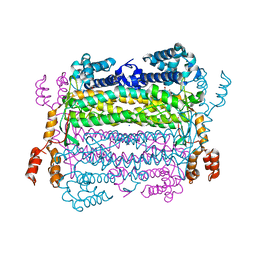 | | Crystal Structure of Adenylosuccinate Lyase from Trypanosoma Brucei, Tb427tmp.160.5560 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Adenylosuccinate lyase, ... | | Authors: | Wernimont, A.K, Loppnau, P, Osman, K.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Robinson, D.A, Wyatt, P.G, Gilbert, I.H, Fairlamb, A.H, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Adenylosuccinate Lyase from Trypanosoma Brucei, Tb427tmp.160.5560
To be Published
|
|
4E5I
 
 | | Crystal structure of avian influenza virus PAn bound to compound 4 | | Descriptor: | 5-hydroxy-2-(1-methyl-1H-imidazol-4-yl)-6-oxo-1,6-dihydropyrimidine-4-carboxylic acid, MANGANESE (II) ION, Polymerase protein PA, ... | | Authors: | DuBois, R.M, Slavish, P.J, Webb, T.R, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.944 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
4EGC
 
 | |
4E83
 
 | |
4ECO
 
 | |
4EJQ
 
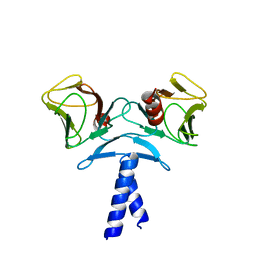 | | Crystal structure of KIF1A C-CC1-FHA | | Descriptor: | Kinesin-like protein KIF1A | | Authors: | Huo, L, Yue, Y, Ren, J, Yu, J, Liu, J, Yu, Y, Ye, F, Xu, T, Zhang, M, Feng, W. | | Deposit date: | 2012-04-06 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | The CC1-FHA Tandem as a Central Hub for Controlling the Dimerization and Activation of Kinesin-3 KIF1A
Structure, 20, 2012
|
|
4EHV
 
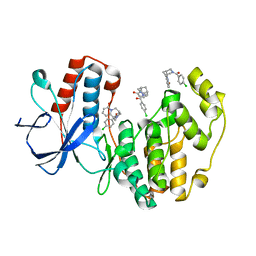 | | Human p38 MAP kinase in complex with NP-F10 and RL87 | | Descriptor: | (1R,5S)-3-(4-chlorobenzoyl)-1,2,3,4,5,6-hexahydro-8H-1,5-methanopyrido[1,2-a][1,5]diazocin-8-one, Mitogen-activated protein kinase 14, N~4~-cyclopropyl-2-phenylquinazoline-4,7-diamine | | Authors: | Over, B, Gruetter, C, Waldmann, H, Rauh, D. | | Deposit date: | 2012-04-04 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Natural-product-derived fragments for fragment-based ligand discovery.
Nat Chem, 5, 2012
|
|
4EI8
 
 | |
4EIF
 
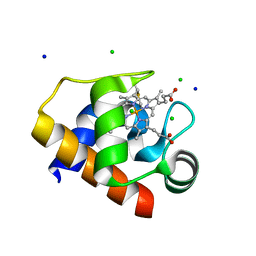 | | Crystal structure of cytochrome c6C L50Q mutant from Synechococcus sp. PCC 7002 | | Descriptor: | CHLORIDE ION, Cytochrome c6, HEME C, ... | | Authors: | Krzywda, S, Bialek, W, Zatwarnicki, P, Jaskolski, M, Szczepaniak, A. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Cytochrome c6 and c6C from Synechococcus sp. PCC 7002 - structure and function.
To be Published
|
|
4E5Q
 
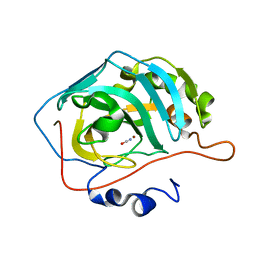 | | Human Carbonic Anhydrase II in complex with cyanate | | Descriptor: | Carbonic anhydrase 2, ZINC ION, cyanic acid | | Authors: | West, D, McKenna, R. | | Deposit date: | 2012-03-14 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7024 Å) | | Cite: | Human carbonic anhydrase II-cyanate inhibitor complex: putting the debate to rest.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4E5V
 
 | |
4F98
 
 | |
4FD0
 
 | |
4FDY
 
 | |
4FJ6
 
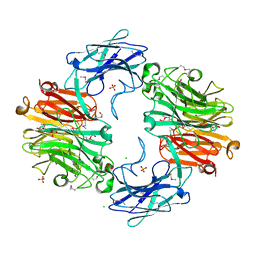 | |
4FMT
 
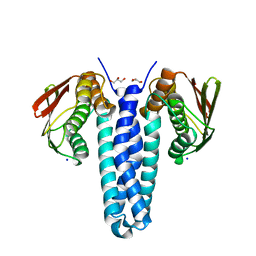 | |
7K53
 
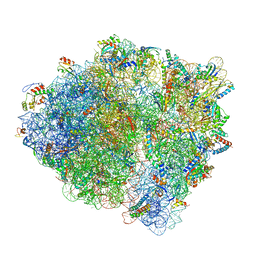 | | Pre-translocation +1-frameshifting(CCC-A) complex (Structure I-FS) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
4FO5
 
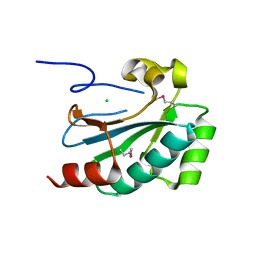 | |
7K51
 
 | | Mid-translocated non-frameshifting(CCA-A) complex with EF-G and GDPCP (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K50
 
 | | Pre-translocation non-frameshifting(CCA-A) complex (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
