7SB2
 
 | |
7SAX
 
 | |
4CKP
 
 | |
4CZH
 
 | | C. crescentus MreB, single filament, ADP, MP265 inhibitor | | Descriptor: | 4-chlorobenzyl carbamimidothioate, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lowe, J, vandenEnt, F. | | Deposit date: | 2014-04-19 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.644 Å) | | Cite: | Bacterial Actin Mreb Forms Antiparallel Double Filaments.
Elife, 3, 2014
|
|
4CZI
 
 | |
4CZL
 
 | | C. crescentus MreB, monomeric, ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ROD SHAPE-DETERMINING PROTEIN MREB | | Authors: | Lowe, J, van den Ent, F. | | Deposit date: | 2014-04-19 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bacterial Actin Mreb Forms Antiparallel Double Filaments.
Elife, 3, 2014
|
|
4CKN
 
 | |
4CZG
 
 | | C. crescentus MreB, single filament, ADP, A22 inhibitor | | Descriptor: | 3,4-dichlorobenzyl carbamimidothioate, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lowe, J, van den Ent, F. | | Deposit date: | 2014-04-19 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bacterial Actin Mreb Forms Antiparallel Double Filaments.
Elife, 3, 2014
|
|
4CSA
 
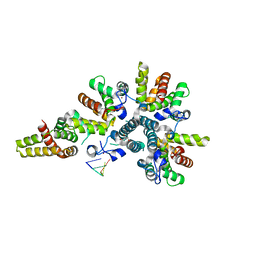 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer bound to a DNA 4-mer | | Descriptor: | 5'-D(*AP*GP*TP*TP*AP)-3', GLYCEROL, M2-1, ... | | Authors: | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
4CZK
 
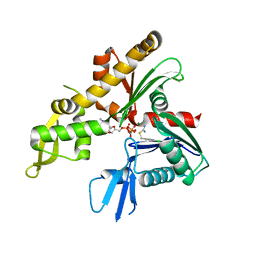 | | C. crescentus MreB, single filament, AMPPNP, MP265 inhibitor | | Descriptor: | 4-chlorobenzyl carbamimidothioate, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Lowe, J, van den Ent, F. | | Deposit date: | 2014-04-19 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Bacterial Actin Mreb Forms Antiparallel Double Filaments.
Elife, 3, 2014
|
|
4CZE
 
 | |
4CZJ
 
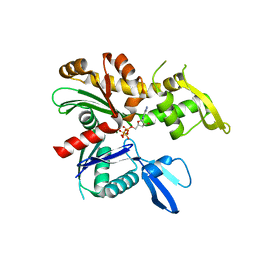 | | C. crescentus MreB, double filament, AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ROD SHAPE-DETERMINING PROTEIN MREB | | Authors: | Lowe, J, van den Ent, F. | | Deposit date: | 2014-04-19 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacterial Actin Mreb Forms Antiparallel Double Filaments.
Elife, 3, 2014
|
|
4CZF
 
 | | C. crescentus MreB, single filament, ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ROD SHAPE-DETERMINING PROTEIN MREB | | Authors: | Lowe, J, vandenEnt, F. | | Deposit date: | 2014-04-19 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Bacterial Actin Mreb Forms Antiparallel Double Filaments.
Elife, 3, 2014
|
|
4CIU
 
 | | Crystal structure of E. coli ClpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHAPERONE PROTEIN CLPB | | Authors: | Kopp, J, Sinning, I, Bukau, B, Kummer, E, Mogk, A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Cooperation with Hsp70 in Protein Disaggregation
Elife, 3, 2014
|
|
7T8W
 
 | | Structure of antibody 3G12 bound to Respiratory Syncytial Virus G central conserved domain mutant S177Q | | Descriptor: | 3G12 Fab Heavy chain, 3G12 Fab Light chain, Mature secreted glycoprotein G | | Authors: | Nunez Castrejon, A.M, O'Rourke, S.M, Kauvar, L.M, DuBois, R.M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Design and Antigenic Validation of Respiratory Syncytial Virus G Immunogens.
J.Virol., 96, 2022
|
|
8AON
 
 | |
6THE
 
 | | Crystal structure of core domain of four-domain heme-cupredoxin-Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-20 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
6FHP
 
 | | DAIP in complex with a C-terminal fragment of thermolysin | | Descriptor: | Dispase autolysis-inducing protein, Thermolysin | | Authors: | Schmelz, S, Fiebig, D, Fuchsbauer, H.L, Blankenfeldt, W, Scrima, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Destructive twisting of neutral metalloproteases: the catalysis mechanism of the Dispase autolysis-inducing protein from Streptomyces mobaraensis DSM 40487.
FEBS J., 285, 2018
|
|
7JH4
 
 | | Crystal structure of NAD(P)H-flavin oxidoreductase (NfoR) from S. aureus complexed with reduced FMN and NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NAD(P)H-dependent oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zheng, Y, O'Neill, A.G, Beaupre, B.A, Liu, D, Moran, G.R. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | NfoR: Chromate Reductase or Flavin Mononucleotide Reductase?
Appl.Environ.Microbiol., 86, 2020
|
|
7K82
 
 | | The X-ray crystal structure of SSR4, an S. pombe chromatin remodelling protein: sulfur SAD | | Descriptor: | CHLORIDE ION, GLYCEROL, SWI/SNF and RSC complexes subunit ssr4 | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X-ray crystal structure of the N-terminal domain of Ssr4, a Schizosaccharomyces pombe chromatin-remodelling protein.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6LCE
 
 | |
6LCF
 
 | | Crystal Structure of beta-L-arabinobiose binding protein - native | | Descriptor: | ABC transporter substrate binding component, beta-L-arabinofuranose-(1-2)-beta-L-arabinofuranose | | Authors: | Miyake, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2019-11-18 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural analysis of beta-L-arabinobiose-binding protein in the metabolic pathway of hydroxyproline-rich glycoproteins in Bifidobacterium longum.
Febs J., 287, 2020
|
|
6L0L
 
 | | Hydra-1ubq de nova designed by Hydra based on ubiquitin | | Descriptor: | Hydra-1ubq | | Authors: | Ouyang, B. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multiobjective heuristic algorithm for de novo protein design in a quantified continuous sequence space.
Comput Struct Biotechnol J, 19, 2021
|
|
4UON
 
 | |
4WA2
 
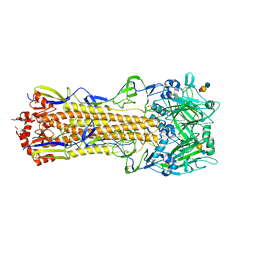 | | The crystal structure of hemagglutinin from a H3N8 influenza virus isolated from New England harbor seals in complex with 3'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Yang, H, Villanueva, J.M, Gubareva, L.V, Stevens, J. | | Deposit date: | 2014-08-28 | | Release date: | 2015-01-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Analysis of Surface Proteins from an A(H3N8) Influenza Virus Isolated from New England Harbor Seals.
J.Virol., 89, 2015
|
|
