5VHI
 
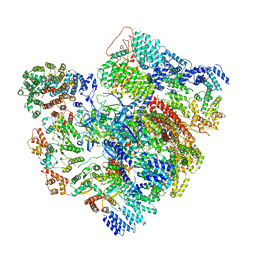 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 10, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
6WQH
 
 | | Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ig2 substrate, Lon protease, ... | | Authors: | Zhang, K, Li, S, Hsiehb, K, Sub, S, Pintilie, G, Chiu, W, Chang, C. | | Deposit date: | 2020-04-28 | | Release date: | 2021-06-09 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis for ATPase-powered substrate translocation by the Lon AAA+ protease.
J.Biol.Chem., 297, 2021
|
|
5WVI
 
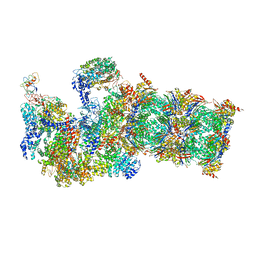 | | The resting state of yeast proteasome | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Ding, Z, Cong, Y. | | Deposit date: | 2016-12-25 | | Release date: | 2017-03-22 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
4HSE
 
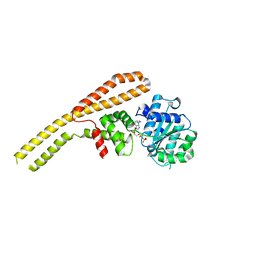 | | Crystal structure of ClpB NBD1 in complex with guanidinium chloride and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Chaperone protein ClpB, ... | | Authors: | Zeymer, C, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular mechanism of hsp100 chaperone inhibition by the prion curing agent guanidinium chloride.
J.Biol.Chem., 288, 2013
|
|
5C1B
 
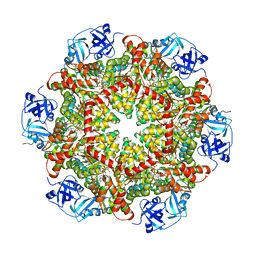 | |
5C1A
 
 | | p97-N750D/R753D/M757D/Q760D in complex with ATP-gamma-S | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2015-06-13 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis of ATP Hydrolysis and Intersubunit Signaling in the AAA+ ATPase p97.
Structure, 24, 2016
|
|
3SYL
 
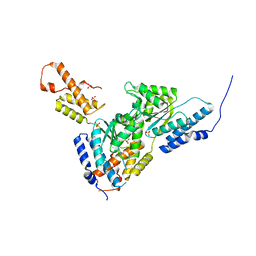 | | Crystal structure of the AAA+ protein CbbX, native structure | | Descriptor: | Protein CbbX, SULFATE ION | | Authors: | Mueller-Cajar, O, Stotz, M, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-18 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase.
Nature, 479, 2011
|
|
3T15
 
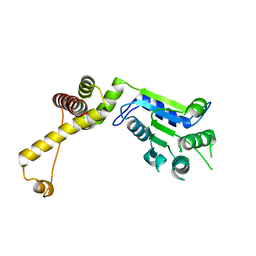 | | Structure of green-type Rubisco activase from tobacco | | Descriptor: | Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplastic | | Authors: | Stotz, M, Wendler, P, Mueller-Cajar, O, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-21 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of green-type Rubisco activase from tobacco.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3SYK
 
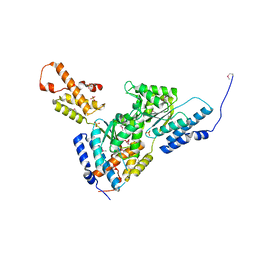 | | Crystal structure of the AAA+ protein CbbX, selenomethionine structure | | Descriptor: | Protein CbbX, SULFATE ION | | Authors: | Mueller-Cajar, O, Stotz, M, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-18 | | Release date: | 2011-11-09 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase.
Nature, 479, 2011
|
|
6WJD
 
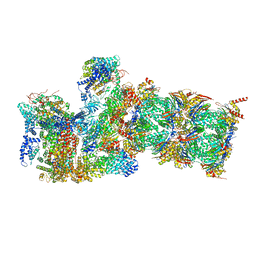 | |
5D4W
 
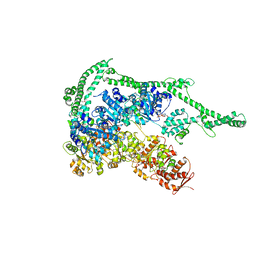 | | Crystal structure of Hsp104 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative heat shock protein | | Authors: | Heuck, A, Schitter-Sollner, S, Clausen, T. | | Deposit date: | 2015-08-09 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the disaggregase activity and regulation of Hsp104.
Elife, 5, 2016
|
|
6WGG
 
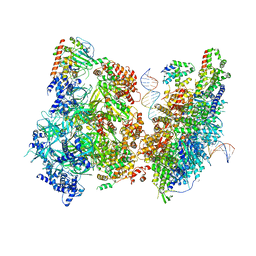 | | Atomic model of pre-insertion mutant OCCM-DNA complex(ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (41-MER), ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WGC
 
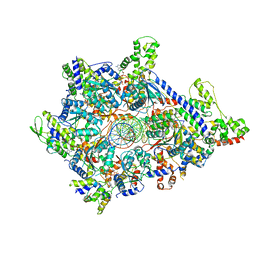 | | Atomic model of semi-attached mutant OCCM-DNA complex (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) | | Descriptor: | Cell division control protein 6, DNA (41-MER), DNA replication licensing factor MCM3, ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4KO8
 
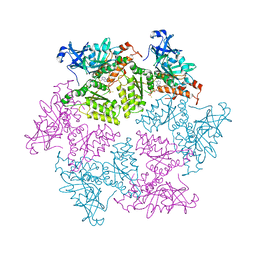 | | Structure of p97 N-D1 R155H mutant in complex with ATPgS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2013-05-11 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Altered Intersubunit Communication Is the Molecular Basis for Functional Defects of Pathogenic p97 Mutants.
J.Biol.Chem., 288, 2013
|
|
4KOD
 
 | | Structure of p97 N-D1 R155H mutant in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2013-05-11 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Altered Intersubunit Communication Is the Molecular Basis for Functional Defects of Pathogenic p97 Mutants.
J.Biol.Chem., 288, 2013
|
|
4KLN
 
 | | Structure of p97 N-D1 A232E mutant in complex with ATPgS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2013-05-07 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Altered Intersubunit Communication Is the Molecular Basis for Functional Defects of Pathogenic p97 Mutants.
J.Biol.Chem., 288, 2013
|
|
6Z1E
 
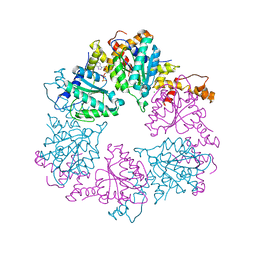 | |
6Z1D
 
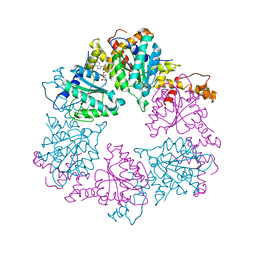 | |
6Z1F
 
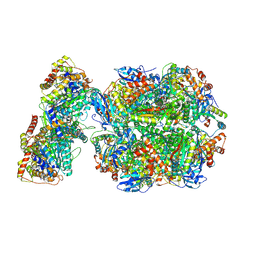 | | CryoEM structure of Rubisco Activase with its substrate Rubisco from Nostoc sp. (strain PCC7120) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, H, Bracher, A, Flecken, M, Popilka, L, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-23 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Dual Functions of a Rubisco Activase in Metabolic Repair and Recruitment to Carboxysomes.
Cell, 183, 2020
|
|
3U5Z
 
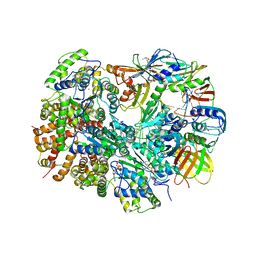 | | Structure of T4 Bacteriophage clamp loader bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase accessory protein 44, DNA polymerase accessory protein 62, ... | | Authors: | Kelch, B.A, Makino, D.L, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2011-10-11 | | Release date: | 2012-01-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | How a DNA polymerase clamp loader opens a sliding clamp.
Science, 334, 2011
|
|
5DYG
 
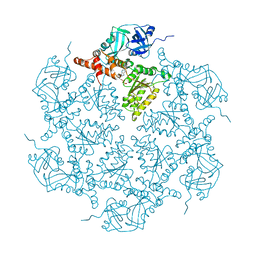 | | Structure of p97 N-D1 L198W mutant in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Tang, W.K, Xia, D. | | Deposit date: | 2015-09-24 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of the D1-D2 Linker of Human VCP/p97 in the Asymmetry and ATPase Activity of the D1-domain.
Sci Rep, 6, 2016
|
|
5C18
 
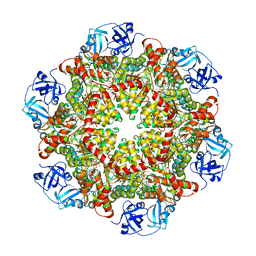 | | p97-delta709-728 in complex with ATP-gamma-S | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2015-06-13 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of ATP Hydrolysis and Intersubunit Signaling in the AAA+ ATPase p97.
Structure, 24, 2016
|
|
5E7P
 
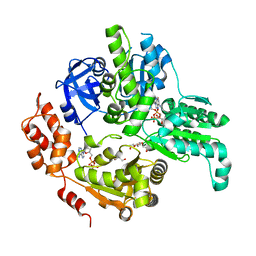 | | Crystal Structure of MSMEG_0858 (Uniprot A0QQS4), a AAA ATPase. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein Cdc48, GLYCEROL, ... | | Authors: | Unciulac-Carp, M, Smith, P, Shuman, S. | | Deposit date: | 2015-10-12 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Crystal Structure and Biochemical Characterization of a Mycobacterium smegmatis AAA-Type Nucleoside Triphosphatase Phosphohydrolase (Msm0858).
J.Bacteriol., 198, 2016
|
|
4L15
 
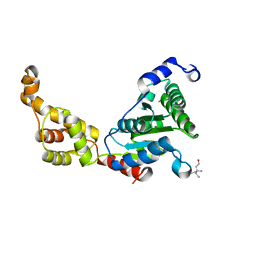 | | Crystal structure of FIGL-1 AAA domain | | Descriptor: | Fidgetin-like protein 1, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Peng, W, Lin, Z, Li, W, Lu, J, Shen, Y, Wang, C. | | Deposit date: | 2013-06-02 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the unusually strong ATPase activity of the AAA domain of the Caenorhabditis elegans fidgetin-like 1 (FIGL-1) protein.
J.Biol.Chem., 288, 2013
|
|
3VFD
 
 | | Human spastin AAA domain | | Descriptor: | SULFATE ION, Spastin | | Authors: | Taylor, J.L, White, S.R, Lauring, B, Kull, F.J. | | Deposit date: | 2012-01-09 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Crystal structure of the human spastin AAA domain.
J.Struct.Biol., 179, 2012
|
|
