5W3Q
 
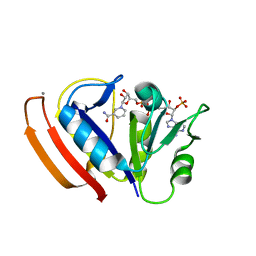 | | L28F E.coli DHFR in complex with NADPH | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Oyen, D, Wright, P.E, Wilson, I.A. | | Deposit date: | 2017-06-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Defining the Structural Basis for Allosteric Product Release from E. coli Dihydrofolate Reductase Using NMR Relaxation Dispersion.
J. Am. Chem. Soc., 139, 2017
|
|
4V66
 
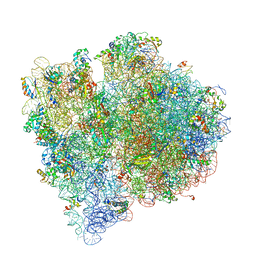 | | Structure of the E. coli ribosome and the tRNAs in Post-accommodation state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Devkota, B, Caulfield, T.R, Tan, R.-Z, Harvey, S.C. | | Deposit date: | 2008-08-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | The Structure of the E. coli Ribosome Before and After Accommodation: Implications for Proofreading
To be Published
|
|
4L8M
 
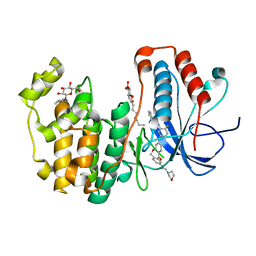 | | Human p38 MAP kinase in complex with a Dibenzoxepinone | | Descriptor: | Mitogen-activated protein kinase 14, N-[2-fluoro-5-({9-[2-(morpholin-4-yl)ethoxy]-11-oxo-6,11-dihydrodibenzo[b,e]oxepin-3-yl}amino)phenyl]benzamide, octyl beta-D-glucopyranoside | | Authors: | Richters, A, Mayer-Wrangowski, S.C, Gruetter, C, Rauh, D. | | Deposit date: | 2013-06-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metabolically Stable Dibenzo[b,e]oxepin-11(6H)-ones as Highly Selective p38 MAP Kinase Inhibitors: Optimizing Anti-Cytokine Activity in Human Whole Blood.
J.Med.Chem., 56, 2013
|
|
4CSY
 
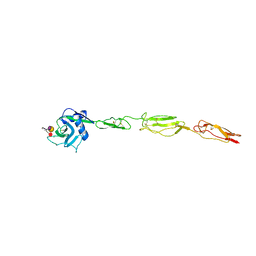 | | E-selectin lectin, EGF-like and two SCR domains complexed with Sialyl Lewis X | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, E-SELECTIN, ... | | Authors: | Preston, R.C, Jakob, R.P, Binder, F.P.C, Sager, C.P, Ernst, B, Maier, T. | | Deposit date: | 2014-03-11 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | E-Selectin Ligand Complexes Adopt an Extended High-Affinity Conformation.
J.Mol.Cell.Biol., 8, 2016
|
|
4UMA
 
 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3 deoxy D arabino heptulosonate 7 phosphate synthase the importance of accommodating the active site water | | Descriptor: | (E)-2-METHYL-3-PHOSPHONOACRYLATE, MANGANESE (II) ION, PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
4L1T
 
 | |
1EOQ
 
 | | ROUS SARCOMA VIRUS CAPSID PROTEIN: C-TERMINAL DOMAIN | | Descriptor: | GAG POLYPROTEIN CAPSID PROTEIN P27 | | Authors: | Kingston, R.L, Fitzon-Ostendorp, T, Eisenmesser, E.Z, Schatz, G.W, Vogt, V.M, Post, C.B, Rossmann, M.G. | | Deposit date: | 2000-03-23 | | Release date: | 2000-08-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and self-association of the Rous sarcoma virus capsid protein.
Structure Fold.Des., 8, 2000
|
|
4U7Y
 
 | | Structure of the complex of VPS4B MIT and IST1 MIM | | Descriptor: | IST1 homolog, Vacuolar protein sorting-associated protein 4B | | Authors: | Guo, E.Z, Xu, Z. | | Deposit date: | 2014-07-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Distinct Mechanisms of Recognizing Endosomal Sorting Complex Required for Transport III (ESCRT-III) Protein IST1 by Different Microtubule Interacting and Trafficking (MIT) Domains.
J.Biol.Chem., 290, 2015
|
|
5W1W
 
 | | Structure of the HLA-E-VMAPRTLVL/GF4 TCR complex | | Descriptor: | Beta-2-microglobulin, GF4 T cell receptor alpha chain, GF4 T cell receptor beta chain, ... | | Authors: | Gras, S, Walpole, N, Farenc, C, Rossjohn, J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A conserved energetic footprint underpins recognition of human leukocyte antigen-E by two distinct alpha beta T cell receptors.
J. Biol. Chem., 292, 2017
|
|
8RLT
 
 | |
8RLV
 
 | |
8RLU
 
 | |
9FNS
 
 | |
5KSR
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-TB (Tetramer Bigger). | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, De Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5KSQ
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa | | Descriptor: | 5'-nucleotidase SurE, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5KST
 
 | | Stationary phase Survival protein E (SurE) from Xylella fastidiosa- XfSurE-TSAmp (Tetramer Smaller - crystallization with 3'AMP). | | Descriptor: | 5'-nucleotidase SurE, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A.S, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.759 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5L4O
 
 | |
4ZT1
 
 | | Crystal structure of human E-Cadherin (residues 3-213) in x-dimer conformation | | Descriptor: | CALCIUM ION, Cadherin-1 | | Authors: | Nardone, V, Lucarelli, A.P, Dalle Vedove, A, Parisini, E. | | Deposit date: | 2015-05-14 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Human E-Cadherin-EC1EC2 in Complex with a Peptidomimetic Competitive Inhibitor of Cadherin Homophilic Interaction.
J.Med.Chem., 59, 2016
|
|
4ZTE
 
 | | Crystal structure of human E-Cadherin (residues 3-213) in complex with a peptidomimetic inhibitor | | Descriptor: | CALCIUM ION, Cadherin-1, N-{[(2S,5S)-1-benzyl-5-(2-{[(2S,3S)-1-(tert-butylamino)-3-methyl-1-oxopentan-2-yl]amino}-2-oxoethyl)-3,6-dioxopiperazin-2-yl]methyl}-L-alpha-asparagine | | Authors: | Nardone, V, Lucarelli, A.P, Dalle Vedove, A, Parisini, E. | | Deposit date: | 2015-05-14 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of Human E-Cadherin-EC1EC2 in Complex with a Peptidomimetic Competitive Inhibitor of Cadherin Homophilic Interaction.
J.Med.Chem., 59, 2016
|
|
5NO6
 
 | | TEAD4-HOXB13 complex bound to DNA | | Descriptor: | DNA, Homeobox protein Hox-B13, Transcriptional enhancer factor TEF-3 | | Authors: | Morgunova, E, Jolma, A, Yin, Y, Popov, A, Taipale, J. | | Deposit date: | 2017-04-11 | | Release date: | 2018-05-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | DNA-guided transcription factor interactions extend human gene regulatory code.
Nature, 2025
|
|
1Y4L
 
 | | Crystal structure of Bothrops asper myotoxin II complexed with the anti-trypanosomal drug suramin | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Murakami, M.T, Arruda, E.Z, Melo, P.A, Martinez, A.B, Calil-Elias, S, Tomaz, M.A, Lomonte, B, Gutierrez, J.M, Arni, R.K. | | Deposit date: | 2004-12-01 | | Release date: | 2005-06-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Myotoxic Activity of Bothrops asper Myotoxin II by the Anti-trypanosomal Drug Suramin.
J.Mol.Biol., 350, 2005
|
|
3TTO
 
 | | Crystal structure of Leuconostoc mesenteroides NRRL B-1299 N-terminally truncated dextransucrase DSR-E in triclinic form | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL | | Authors: | Brison, Y, Pijning, T, Fabre, E, Mourey, L, Morel, S, Potocki-Veronese, G, Monsan, P, Tranier, S, Remaud-Simeon, M, Dijkstra, B.W. | | Deposit date: | 2011-09-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Functional and structural characterization of alpha-(1-2) branching sucrase derived from DSR-E glucansucrase
J.Biol.Chem., 287, 2012
|
|
2VMK
 
 | | Crystal Structure of E. coli RNase E Apoprotein - Catalytic Domain | | Descriptor: | RIBONUCLEASE E, SULFATE ION, ZINC ION | | Authors: | Koslover, D.J, Callaghan, A.J, Marcaida, M.J, Martick, M, Scott, W.G, Luisi, B.F. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of the Escherichia Coli Rnase E Apoprotein and a Mechanism for RNA Degradation.
Structure, 16, 2008
|
|
6FK0
 
 | |
5UOY
 
 | | Crystal structure of human PDE1B catalytic domain in complex with inhibitor 16j (6-(4-Methoxybenzyl)-9-((tetrahydro-2H-pyran-4-yl)methyl)-8,9,10,11-tetrahydropyrido[4',3':4,5]thieno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one) | | Descriptor: | 6-[(4-methoxyphenyl)methyl]-9-[(oxan-4-yl)methyl]-8,9,10,11-tetrahydropyrido[4',3':4,5]thieno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, MAGNESIUM ION, ... | | Authors: | Cedervall, E.P, Allerston, C.K, Xu, R, Sridhar, V, Barker, R, Aertgeerts, K. | | Deposit date: | 2017-02-01 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of Selective Phosphodiesterase 1 Inhibitors with Memory Enhancing Properties.
J. Med. Chem., 60, 2017
|
|
