4UCV
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-methoxy-2,3-dimethylquinoxalin-5-ol, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCR
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-hydroxy-2-methylquinoline-6-carboxamide, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCO
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 7-amino-2-tert-butyl-4-(1H-pyrrol-2-yl)pyrido[2,3-d]pyrimidine-6-carboxamide, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCS
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 5-amino-3-(furan-2-yl)-1H-1,2,4-triazole-1-carboxamide, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
7K72
 
 | |
3SGI
 
 | |
4LH7
 
 | | Crystal structure of a LigA inhibitor | | Descriptor: | 4-aminothieno[3,2-c]pyridine-2,7-dicarboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Boriack-Sjodin, P.A, Prince, D.B. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification through structure-based methods of a bacterial NAD(+)-dependent DNA ligase inhibitor that avoids known resistance mutations.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4LH6
 
 | | Crystal structure of a LigA inhibitor | | Descriptor: | 4-amino-2-bromothieno[3,2-c]pyridine-7-carboxamide, ACETATE ION, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, ... | | Authors: | Benenato, K, Wang, H, Mcguire, H.M, Davis, H, Gao, N, Prince, D.B, Jahic, H, Stokes, S.S, Boriack-Sjodin, P.A. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification through structure-based methods of a bacterial NAD(+)-dependent DNA ligase inhibitor that avoids known resistance mutations.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4EEQ
 
 | | Crystal structure of E. faecalis DNA ligase with inhibitor | | Descriptor: | 4-amino-2-(cyclopentyloxy)pyrimidine-5-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Wang, T, Charifson, P, Wei, Y. | | Deposit date: | 2012-03-28 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis and Activity Evaluation of Potent NAD+ DNA Ligase Inhibitors as Potential Antibacterial Agents.
To be Published
|
|
4EFE
 
 | | crystal structure of DNA ligase | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, SULFATE ION, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of DNA ligase
To be Published
|
|
4EFB
 
 | | Crystal structure of DNA ligase | | Descriptor: | 4-amino-2-(cyclopentyloxy)-6-{[(1R,2S)-2-hydroxycyclopentyl]oxy}pyrimidine-5-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of DNA ligase
To be Published
|
|
3BA8
 
 | | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase | | Descriptor: | 2-amino-7-fluoro-5-oxo-5H-chromeno[2,3-b]pyridine-3-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Pinko, C. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase
To be Published
|
|
3BAA
 
 | | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase | | Descriptor: | 7-amino-2-tert-butyl-4-{[2-(1H-imidazol-4-yl)ethyl]amino}pyrido[2,3-d]pyrimidine-6-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Pinko, C. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase
To be Published
|
|
3BAB
 
 | | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase | | Descriptor: | 7-amino-2-tert-butyl-4-(4-pyrimidin-2-ylpiperazin-1-yl)pyrido[2,3-d]pyrimidine-6-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Pinko, C. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase
To be Published
|
|
3QWU
 
 | | Putative ATP-dependent DNA ligase from Aquifex aeolicus. | | Descriptor: | ADENOSINE, CALCIUM ION, DNA ligase, ... | | Authors: | Osipiuk, J, Quartey, P, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-28 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Putative ATP-dependent DNA ligase from Aquifex aeolicus.
To be Published
|
|
5OP0
 
 | |
6BKG
 
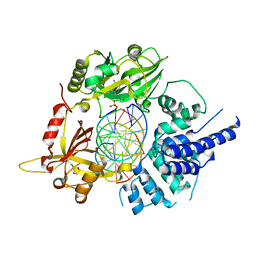 | | Human LigIV catalytic domain with bound DNA-adenylate intermediate in closed conformation | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Moon, A.F, Tumbale, P.P, Schellenberg, M.J, Williams, R.S, Williams, J.G, Kunkel, T.A, Pedersen, L.C, Bebenek, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structures of DNA-bound human ligase IV catalytic core reveal insights into substrate binding and catalysis.
Nat Commun, 9, 2018
|
|
6BKF
 
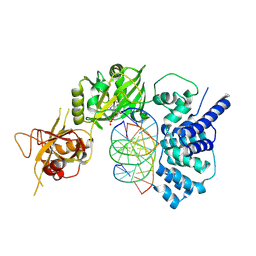 | | Lysyl-adenylate form of human LigIV catalytic domain with bound DNA substrate in open conformation | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*C)-3'), DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Moon, A.F, Tumbale, P.P, Schellenberg, M.J, Williams, R.S, Williams, J.G, Kunkel, T.A, Pedersen, L.C, Bebenek, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structures of DNA-bound human ligase IV catalytic core reveal insights into substrate binding and catalysis.
Nat Commun, 9, 2018
|
|
4RKI
 
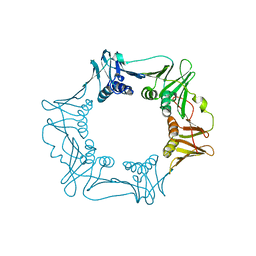 | |
4S3I
 
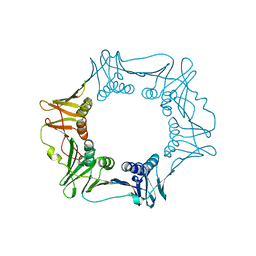 | |
2IRX
 
 | |
3RB9
 
 | |
2IRY
 
 | |
2IRU
 
 | |
7FB7
 
 | | Crystal structure of human UHRF1 TTD in complex with 5-amino-2,4-dimethylpyridine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-amino-2,4-dimethylpyridine, DIMETHYL SULFOXIDE, ... | | Authors: | Kori, S, Arita, K, Yoshimi, S. | | Deposit date: | 2021-07-08 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-based screening combined with computational and biochemical analyses identified the inhibitor targeting the binding of DNA Ligase 1 to UHRF1.
Bioorg.Med.Chem., 52, 2021
|
|
