6T2S
 
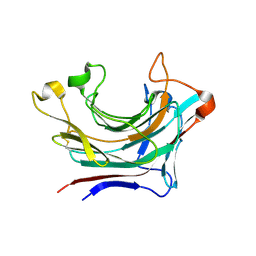 | | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown | | Descriptor: | Glycoside hydrolase family 16 protein, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose | | Authors: | Crouch, L.I, Liberato, M.V, Ubranowicz, P.A, Basle, A, Lamb, C.A, Cooke, K, Doona, M, Needham, S, Brady, R.R, Berrington, J.E, Madubic, K, Chater, P, Zhang, F, Linhardt, R.J, Spence, D.I.R, Bolam, D.N. | | Deposit date: | 2019-10-09 | | Release date: | 2020-07-08 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown.
Nat Commun, 11, 2020
|
|
8YPU
 
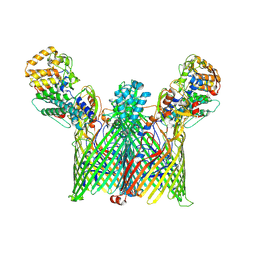 | | Cryo-EM structure of ButCD complex | | Descriptor: | Membrane protein, SusC/RagA family TonB-linked outer membrane protein | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2024-03-18 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | A highly conserved SusCD transporter determines the import and species-specific antagonism of Bacteroides ubiquitin homologues
Nat Commun, 2024
|
|
8DGO
 
 | |
8DEB
 
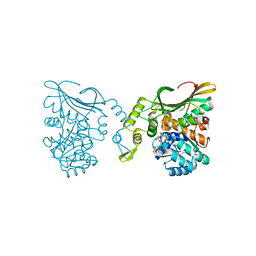 | | Bacteroides fragilis carboxyspermidine dehydrogenase | | Descriptor: | Carboxyspermidine dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | McFarlane, J.S, Bouchey, S, Dodd, J. | | Deposit date: | 2022-06-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Kinetic and structural characterization of carboxyspermidine dehydrogenase of polyamine biosynthesis.
J.Biol.Chem., 299, 2023
|
|
5KNH
 
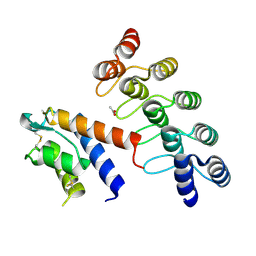 | | CRYSTAL STRUCTURE OF DARPIN 6G9 IN COMPLEX WITH CYNO IL-13 | | Descriptor: | ACETATE ION, DARPIN 6G9, IL13 | | Authors: | Teplyakov, A, Malia, T, Obmolova, G, Gilliland, G. | | Deposit date: | 2016-06-28 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational flexibility of an anti-IL-13 DARPin.
Protein Eng. Des. Sel., 30, 2017
|
|
8FL6
 
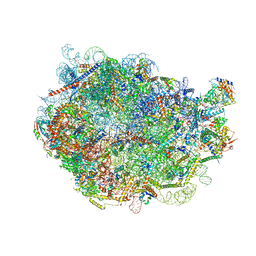 | |
2QZU
 
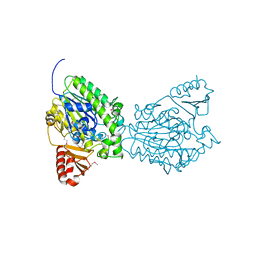 | | Crystal structure of the putative sulfatase yidJ from Bacteroides fragilis. Northeast Structural Genomics Consortium target BfR123 | | Descriptor: | Putative sulfatase yidJ | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Wang, D, Cunningham, K, Maglaqui, M, Owens, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-08-17 | | Release date: | 2007-09-04 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystal structure of the putative sulfatase yidJ from Bacteroides fragilis.
To be Published
|
|
2FG6
 
 | | N-succinyl-L-ornithine transcarbamylase from B. fragilis complexed with sulfate and N-succinyl-L-norvaline | | Descriptor: | N-(3-CARBOXYPROPANOYL)-L-NORVALINE, SULFATE ION, putative ornithine carbamoyltransferase | | Authors: | Shi, D, Yu, X, Malamy, M.H, Allewell, N.M, Mendel, T. | | Deposit date: | 2005-12-21 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and catalytic mechanism of a novel N-succinyl-L-ornithine transcarbamylase in arginine biosynthesis of Bacteroides fragilis.
J.Biol.Chem., 281, 2006
|
|
2QR3
 
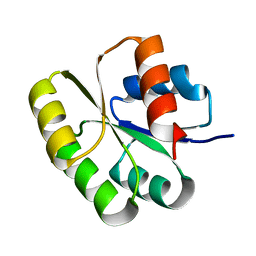 | | Crystal structure of the N-terminal signal receiver domain of two-component system response regulator from Bacteroides fragilis | | Descriptor: | Two-component system response regulator | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Mendoza, M, Romero, R, Fong, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the N-terminal signal receiver domain of two-component system response regulator from Bacteroides fragilis.
To be Published
|
|
6USS
 
 | |
4R37
 
 | | Crystal structure analysis of LpxA, a UDP-N-acetylglucosamine acyltransferase from Bacteroides fragilis 9343 with UDP-GlcNAc | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETATE ION, ... | | Authors: | Fisher, A.J, Chen, X, Ngo, A. | | Deposit date: | 2014-08-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Bacteroides fragilis uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (BfLpxA).
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8RJD
 
 | |
4R36
 
 | | Crystal structure analysis of LpxA, a UDP-N-acetylglucosamine acyltransferase from Bacteroides fragilis 9343 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETATE ION, ... | | Authors: | Ngo, A, Fong, K, Cox, D, Fisher, A, Chen, X. | | Deposit date: | 2014-08-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Bacteroides fragilis uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (BfLpxA).
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RAA
 
 | |
8SWT
 
 | |
5Z1A
 
 | |
6WZK
 
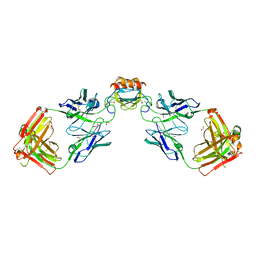 | | LY3041658 Fab bound to CXCL3 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, C-X-C motif chemokine 3, ... | | Authors: | Durbin, J.D, Druzina, Z. | | Deposit date: | 2020-05-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and characterization of a neutralizing pan-ELR+CXC chemokine monoclonal antibody.
Mabs, 12
|
|
4GBS
 
 | |
5YYS
 
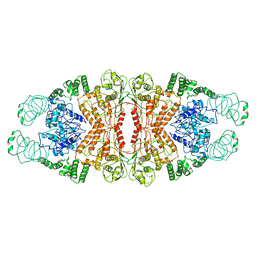 | | Cryo-EM structure of L-fucokinase, GDP-fucose pyrophosphorylase (FKP)in Bacteroides fragilis | | Descriptor: | L-fucokinase, L-fucose-1-P guanylyltransferase | | Authors: | Wang, J, Hu, H, Liu, Y, Zhou, Q, Wu, P, Yan, N, Wang, H.W, Wu, J.W, Sun, L. | | Deposit date: | 2017-12-11 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of L-fucokinase/GDP-fucose pyrophosphorylase (FKP) in Bacteroides fragilis.
Protein Cell, 10, 2019
|
|
6WZL
 
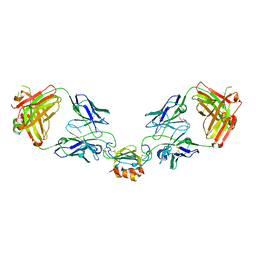 | | LY3041658 Fab bound to CXCL7 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, C-X-C motif chemokine 7, LY3041658 Fab heavy chain, ... | | Authors: | Durbin, J.D, Druzina, Z. | | Deposit date: | 2020-05-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery and characterization of a neutralizing pan-ELR+CXC chemokine monoclonal antibody.
Mabs, 12
|
|
6WZJ
 
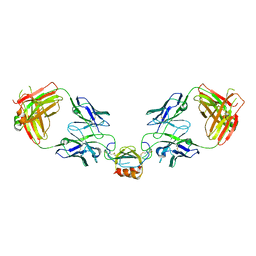 | | LY3041658 Fab bound to CXCL2 | | Descriptor: | C-X-C motif chemokine 2, LY3041658 Fab Heavy Chain, LY3041658 Fab Light Chain | | Authors: | Durbin, J.D, Druzina, Z. | | Deposit date: | 2020-05-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery and characterization of a neutralizing pan-ELR+CXC chemokine monoclonal antibody.
Mabs, 12
|
|
1YIB
 
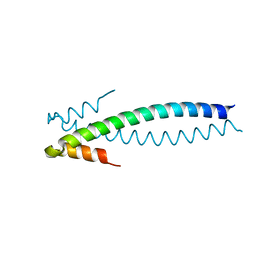 | | Crystal Structure of the Human EB1 C-terminal Dimerization Domain | | Descriptor: | Microtubule-associated protein RP/EB family member 1 | | Authors: | Slep, K.C, Rogers, S.L, Elliott, S.L, Ohkura, H, Kolodziej, P.A, Vale, R.D. | | Deposit date: | 2005-01-11 | | Release date: | 2005-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end
J.Cell Biol., 168, 2005
|
|
4FR9
 
 | |
4H40
 
 | |
2KC7
 
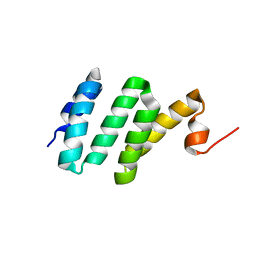 | | Solution NMR structure of Bacteroides fragilis protein BF1650. Northeast Structural Genomics Consortium target BfR218 | | Descriptor: | bfr218_protein | | Authors: | Eletsky, A, Wu, Y, Sukumaran, D, Lee, H, Lee, D.Y, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-17 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Bacteroides fragilis protein BF1650. Northeast Structural Genomics Consortium target BfR218
To be Published
|
|
