2NTH
 
 | | Structure of Spin-labeled T4 Lysozyme Mutant L118R1 | | Descriptor: | Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2006-11-07 | | Release date: | 2007-06-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
1KXV
 
 | | Camelid VHH Domains in Complex with Porcine Pancreatic alpha-Amylase | | Descriptor: | ALPHA-AMYLASE, PANCREATIC, CAMELID VHH DOMAIN CAB10 | | Authors: | Desmyter, A, Spinelli, S, Payan, F, Lauwereys, M, Wyns, L, Muyldermans, S, Cambillau, C. | | Deposit date: | 2002-02-01 | | Release date: | 2002-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology.
J.Biol.Chem., 277, 2002
|
|
1KXQ
 
 | | Camelid VHH Domain in Complex with Porcine Pancreatic alpha-Amylase | | Descriptor: | CALCIUM ION, CHLORIDE ION, alpha-amylase, ... | | Authors: | Desmyter, A, Spinelli, S, Payan, F, Lauwereys, M, Wyns, L, Muyldermans, S, Cambillau, C. | | Deposit date: | 2002-02-01 | | Release date: | 2002-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology.
J.Biol.Chem., 277, 2002
|
|
2I5P
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase isoform 1 from K. marxianus | | Descriptor: | BETA-MERCAPTOETHANOL, Glyceraldehyde-3-phosphate dehydrogenase 1, alpha-D-glucopyranose | | Authors: | Ferreira-da-Silva, F, Pereira, P.J.B, Gales, L, Moradas-Ferreira, P, Damas, A.M. | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal and Solution Structures of Glyceraldehyde-3-phosphate Dehydrogenase Reveal Different Quaternary Structures.
J.Biol.Chem., 281, 2006
|
|
2OU8
 
 | | Structure of Spin-labeled T4 Lysozyme Mutant T115R1 at Room Temperature | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-02-09 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | EPR (1.8 Å), X-RAY DIFFRACTION | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
2OU9
 
 | | Structure of Spin-labeled T4 Lysozyme Mutant T115R1/R119A | | Descriptor: | Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-02-09 | | Release date: | 2007-06-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
2PC5
 
 | | Native crystal structure analysis on Arabidopsis dUTPase | | Descriptor: | DUTP pyrophosphatase-like protein, MAGNESIUM ION | | Authors: | Moriyama, H, Bajaj, M. | | Deposit date: | 2007-03-29 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Purification, crystallization and preliminary crystallographic analysis of deoxyuridine triphosphate nucleotidohydrolase from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2OYP
 
 | | T Cell Immunoglobulin Mucin-3 Crystal Structure Revealed a Galectin-9-independent Binding Surface | | Descriptor: | Hepatitis A virus cellular receptor 2, SULFATE ION | | Authors: | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | T cell immunoglobulin mucin-3 crystal structure reveals a galectin-9-independent ligand-binding surface
Immunity, 26, 2007
|
|
1SQF
 
 | | The crystal structure of E. coli Fmu binary complex with S-Adenosylmethionine at 2.1 A resolution | | Descriptor: | S-ADENOSYLMETHIONINE, SUN protein | | Authors: | Foster, P.G, Nunes, C.R, Greene, P, Moustakas, D, Stroud, R.M. | | Deposit date: | 2004-03-18 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The First Structure of an RNA m5C Methyltransferase,
Fmu, Provides Insight into Catalytic Mechanism
and Specific Binding of RNA Substrate
Structure, 11, 2003
|
|
2GSC
 
 | | Crystal Structure of the Conserved Hypothetical Cytosolic Protein Xcc0516 from Xanthomonas campestris | | Descriptor: | Putative uncharacterized protein XCC0516 | | Authors: | Lin, L.Y, Ching, C.L, Chin, K.H, Chou, S.H, Chan, N.L. | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the conserved hypothetical cytosolic protein Xcc0516 from Xanthomonas campestris reveals a novel quaternary structure assembled by five four-helix bundles.
Proteins, 65, 2006
|
|
2P5Z
 
 | | The E. coli c3393 protein is a component of the type VI secretion system and exhibits structural similarity to T4 bacteriophage tail proteins gp27 and gp5 | | Descriptor: | Type VI secretion system component | | Authors: | Ramagopal, U.A, Bonanno, J.B, Sridhar, V, Lau, C, Toro, R, Gheyi, T, Maletic, M, Freeman, J.C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin.
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
2PLY
 
 | | Structure of the mRNA binding fragment of elongation factor SelB in complex with SECIS RNA. | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Soler, N, Fourmy, D, Yoshizawa, S. | | Deposit date: | 2007-04-20 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into a molecular switch in tandem winged-helix motifs from elongation factor SelB.
J.Mol.Biol., 370, 2007
|
|
2N8Q
 
 | |
2GL2
 
 | | Crystal structure of the tetra mutant (T66G,R67G,F68G,Y69G) of bacterial adhesin FadA | | Descriptor: | adhesion A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2006-04-04 | | Release date: | 2007-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallization and preliminary X-ray data of the FadA adhesin from Fusobacterium nucleatum.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1PJ3
 
 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate pyruvate, cofactor NAD+, Mn++, and allosteric activator fumarate. | | Descriptor: | FUMARIC ACID, MANGANESE (II) ION, NAD-dependent malic enzyme, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-30 | | Release date: | 2003-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
2HB0
 
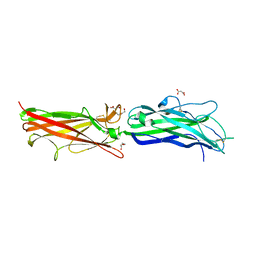 | | Crystal Structure of CfaE, the Adhesive Subunit of CFA/I Fimbria of Enterotoxigenic Escherichia coli | | Descriptor: | CFA/I fimbrial subunit E, DI(HYDROXYETHYL)ETHER, MALONATE ION | | Authors: | Li, Y.F, Xia, D, Poole, S, Rasulova, F, Savarino, S.J. | | Deposit date: | 2006-06-13 | | Release date: | 2007-06-26 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A receptor-binding site as revealed by the crystal structure of CfaE, the colonization factor antigen I fimbrial adhesin of enterotoxigenic Escherichia coli.
J.Biol.Chem., 282, 2007
|
|
2NAU
 
 | |
2NAT
 
 | |
1M5H
 
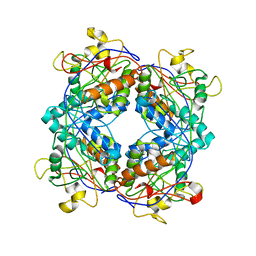 | | Formylmethanofuran:tetrahydromethanopterin formyltransferase from Archaeoglobus fulgidus | | Descriptor: | Formylmethanofuran--tetrahydromethanopterin formyltransferase, POTASSIUM ION | | Authors: | Mamat, B, Roth, A, Grimm, C, Ermler, U, Tziatzios, C, Schubert, D, Thauer, R.K, Shima, S. | | Deposit date: | 2002-07-09 | | Release date: | 2002-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and enzymatic properties of three formyltransferases from archaea: environmental adaptation and evolutionary relationship.
Protein Sci., 11, 2002
|
|
1SV5
 
 | | CRYSTAL STRUCTURE OF K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R165335 | | Descriptor: | 4-({6-AMINO-5-BROMO-2-[(4-CYANOPHENYL)AMINO]PYRIMIDIN-4-YL}OXY)-3,5-DIMETHYLBENZONITRILE, Reverse Transcriptase | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-03-27 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
2MUZ
 
 | | ssNMR structure of a designed rocker protein | | Descriptor: | designed rocker protein | | Authors: | Wang, T, Joh, N, Wu, Y, DeGrado, W.F, Hong, M. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | De novo design of a transmembrane Zn2+-transporting four-helix bundle.
Science, 346, 2014
|
|
2N1Q
 
 | | HIV-1 Core Packaging Signal | | Descriptor: | RNA_(155-MER) | | Authors: | Keane, S.C, Summers, M.F. | | Deposit date: | 2015-04-15 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RNA structure. Structure of the HIV-1 RNA packaging signal.
Science, 348, 2015
|
|
2GLZ
 
 | |
2PLQ
 
 | | Crystal structure of the amidase from geobacillus pallidus RAPc8 | | Descriptor: | Aliphatic amidase | | Authors: | Kimani, S.W, Sewell, B.T, Agarkar, V.B, Sayed, M.F, Cowan, D.A. | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The quaternary structure of the amidase from Geobacillus pallidus RAPc8 is
revealed by its crystal packing.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2N62
 
 | | ddFLN5+110 | | Descriptor: | gelation factor, secretion monitor chimera | | Authors: | Cabrita, L.D, Cassaignau, A.M.E, Launay, H.M.M, Waudby, C.A, Camilloni, C, Robertson, A.L, Wang, X, Wlodarski, T, Wentink, A.S, Vendruscolo, M, Dobson, C.M, Christodoulou, J. | | Deposit date: | 2015-08-10 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding.
Nat.Struct.Mol.Biol., 23, 2016
|
|
