8WOG
 
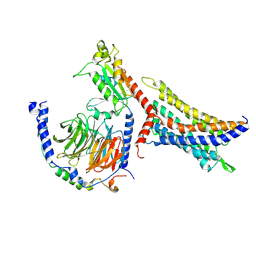 | | Cryo-EM structure of SUCR1 in complex with succinate and Gi protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Liu, A, Ye, R.D. | | Deposit date: | 2023-10-07 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into ligand recognition and activation of the succinate receptor SUCNR1.
Cell Rep, 43, 2024
|
|
8W2H
 
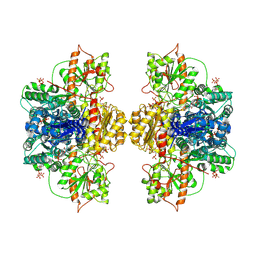 | | Human liver phosphofructokinase-1 in the T-state conformation | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent 6-phosphofructokinase, ... | | Authors: | Lynch, E.M, Kollman, J.M, Webb, B.A. | | Deposit date: | 2024-02-20 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for allosteric regulation of human phosphofructokinase-1.
Nat Commun, 15, 2024
|
|
7CH9
 
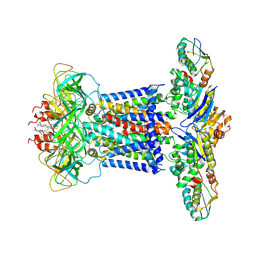 | | Cryo-EM structure of P.aeruginosa MlaFEBD | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, MlaD domain-containing protein, Probable ATP-binding component of ABC transporter, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
4G8M
 
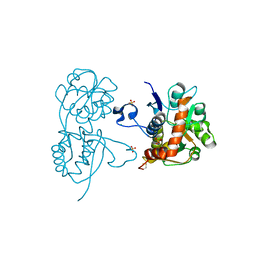 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the agonist CBG-IV at 2.05A resolution | | Descriptor: | (1S,2R)-2-[(S)-amino(carboxy)methyl]cyclobutanecarboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Juknaite, L, Frydenvang, K, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pharmacological and structural characterization of conformationally restricted (S)-glutamate analogues at ionotropic glutamate receptors.
J.Struct.Biol., 180, 2012
|
|
8WRO
 
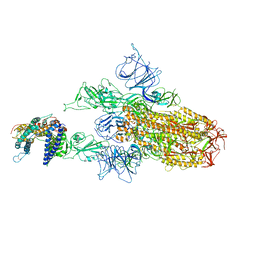 | | XBB.1.5.10 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein,Spike glycoprotein,Spike glycoprotein,Fusion protein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTD
 
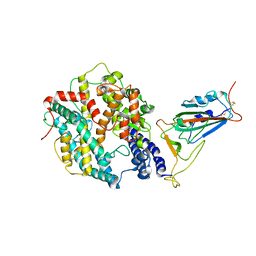 | | XBB.1.5.10 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTJ
 
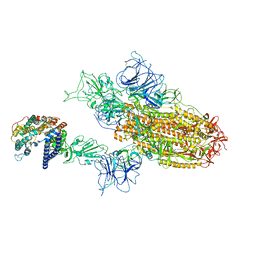 | | XBB.1.5.70 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.64 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
5AIG
 
 | | Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. Tomsk-sample- Valpromide complex | | Descriptor: | 2-PROPYLPENTANAMIDE, DIMETHYL SULFOXIDE, LIMONENE-1,2-EPOXIDE HYDROLASE | | Authors: | Ferrandi, E, Sayer, C, Isupov, M.N, Annovazzi, C, Marchesi, C, Iacobone, G, Peng, X, Bonch-Osmolovskaya, E, Wohlgemuth, R, Littlechild, J.A, Montia, D. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Discovery and Characterization of Thermophilic Limonene-1,2-Epoxide Hydrolases from Hot Spring Metagenomic Libraries
FEBS J., 282, 2015
|
|
5AII
 
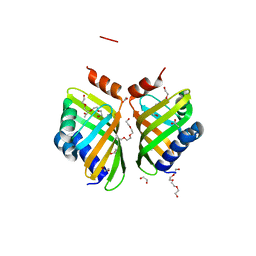 | | Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. CH55-sample-PEG complex | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Ferrandi, E, Sayer, C, Isupov, M.N, Annovazzi, C, Marchesi, C, Iacobone, G, Peng, X, Bonch-Osmolovskaya, E, Wohlgemuth, R, Littlechild, J.A, Montia, D. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery and Characterization of Thermophilic Limonene-1,2-Epoxide Hydrolases from Hot Spring Metagenomic Libraries
FEBS J., 282, 2015
|
|
8A05
 
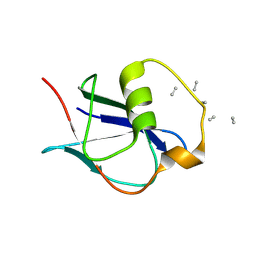 | | Bacteriophage phiCjT23 spike protein penton domain | | Descriptor: | Spike protein P13 N-terminal, capsid internal domain, Unknown vertex protein | | Authors: | Rissanen, I, Huiskonen, J.T. | | Deposit date: | 2022-05-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of ssDNA bacteriophage Phi CjT23 provides insight into early virus evolution.
Nat Commun, 13, 2022
|
|
8A02
 
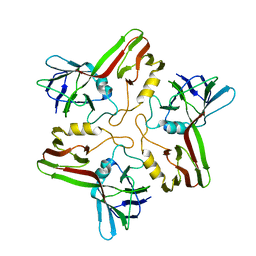 | |
8A06
 
 | |
7ZZZ
 
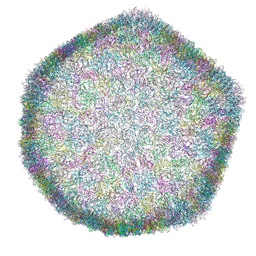 | | Bacteriophage phiCjT23 capsid | | Descriptor: | Major capsid protein P5, Spike protein P13 N-terminal, capsid internal domain, ... | | Authors: | Rissanen, I, Kejzar, N, Huiskonen, J.T. | | Deposit date: | 2022-05-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of ssDNA bacteriophage Phi CjT23 provides insight into early virus evolution.
Nat Commun, 13, 2022
|
|
8A01
 
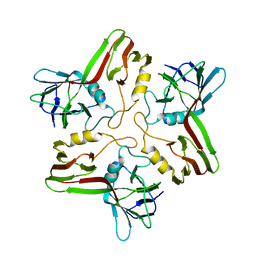 | |
8A04
 
 | |
8A03
 
 | |
5AIH
 
 | | Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. CH55-sample-Native | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, LIMONENE-1,2-EPOXIDE HYDROLASE | | Authors: | Ferrandi, E, Sayer, C, Isupov, M.N, Annovazzi, C, Marchesi, C, Iacobone, G, Peng, X, Bonch-Osmolovskaya, E, Wohlgemuth, R, Littlechild, J.A, Montia, D. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery and Characterization of Thermophilic Limonene-1,2-Epoxide Hydrolases from Hot Spring Metagenomic Libraries
FEBS J., 282, 2015
|
|
7ZF1
 
 | | Structure of ubiquitinated FANCI in complex with FANCD2 and double-stranded DNA | | Descriptor: | DNA (61-MER), Fanconi anemia group D2 protein, Fanconi anemia group I protein, ... | | Authors: | Lemonidis, K, Rennie, M.L, Arkinson, C, Streetley, J, Clarke, M, Chaugule, V.K, Walden, H. | | Deposit date: | 2022-03-31 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structural and biochemical basis of interdependent FANCI-FANCD2 ubiquitination.
Embo J., 42, 2023
|
|
8V4Q
 
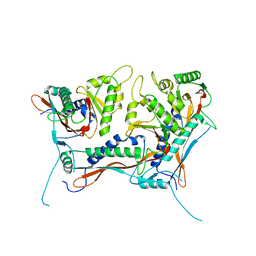 | | Myxococcus xanthus EncA 3xHis pore mutant with tetrahedral symmetry | | Descriptor: | Type 1 encapsulin shell protein EncA | | Authors: | Szyszka, T.N, Andreas, M.P, Lie, F, Miller, L.M, Adamson, L.S.R, Fatehi, F, Twarock, R, Draper, B.E, Jarrold, M.F, Giessen, T.W, Lau, Y.H. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Point mutation in a virus-like capsid drives symmetry reduction to form tetrahedral cages.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6BOA
 
 | | Cryo-EM structure of human TRPV6-R470E in amphipols | | Descriptor: | Transient receptor potential cation channel subfamily V member 6 | | Authors: | McGoldrick, L.L, Singh, A.K, Saotome, K, Yelshanskaya, M.V, Twomey, E.C, Grassucci, R.A, Sobolevsky, A.I. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Opening of the human epithelial calcium channel TRPV6.
Nature, 553, 2018
|
|
6BO9
 
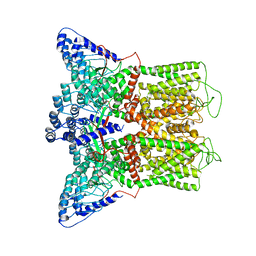 | | Cryo-EM structure of human TRPV6 in amphipols | | Descriptor: | Transient receptor potential cation channel subfamily V member 6 | | Authors: | McGoldrick, L.L, Singh, A.K, Saotome, K, Yelshanskaya, M.V, Twomey, E.C, Grassucci, R.A, Sobolevsky, A.I. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Opening of the human epithelial calcium channel TRPV6.
Nature, 553, 2018
|
|
6HKC
 
 | |
6WU3
 
 | |
8PSL
 
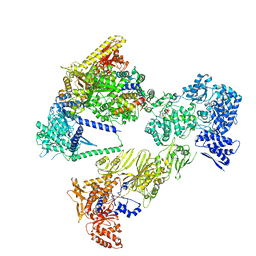 | | Asymmetric unit of the yeast fatty acid synthase in the semi non-rotated state with ACP at the ketosynthase domain (FASx sample) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PS2
 
 | | Asymmetric unit of the yeast fatty acid synthase with ACP at the enoyl reductase domain (FASam sample) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta, ... | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
