2PDI
 
 | | Human aldose reductase mutant L300A complexed with zopolrestat at 1.55 A. | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-04-01 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
2PDY
 
 | | Human aldose reductase double mutant S302R-C303D complexed with fidarestat. | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-04-01 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
2PD9
 
 | | Human aldose reductase mutant V47I complexed with fidarestat. | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-03-31 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
2PDF
 
 | | Human aldose reductase mutant L300P complexed with zopolrestat. | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-03-31 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
1PH1
 
 | |
1PH2
 
 | |
1PH3
 
 | |
1PH4
 
 | |
1PH5
 
 | |
1PH8
 
 | |
1PH6
 
 | |
1PHJ
 
 | |
1PH9
 
 | |
2C2T
 
 | | Human Dihydrofolate Reductase Complexed With NADPH and 2,4-Diamino-5-((7,8-dicarbaundecaboran-7-yl)methyl)-6-methylpyrimidine, a novel boron containing, nonclassical Antifolate | | Descriptor: | (S)-2,4-DIAMINO-5-((7,8-DICARBAUNDECABORAN-7-YL)METHYL)-6-METHYLPYRIMIDINE, DIHYDROFOLATE REDUCTASE, GLYCEROL, ... | | Authors: | leung, A.K.W, reynolds, R.C, riordan, J.M, borhani, D.W. | | Deposit date: | 2005-09-30 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel Boron-Containing, Nonclassical Antifolates: Synthesis and Preliminary Biological and Structural Evaluation.
J.Med.Chem., 50, 2007
|
|
1M3V
 
 | | FLIN4: Fusion of the LIM binding domain of Ldb1 and the N-terminal LIM domain of LMO4 | | Descriptor: | ZINC ION, fusion of the LIM interacting domain of ldb1 and the N-terminal LIM domain of LMO4 | | Authors: | Deane, J.E, Mackay, J.P, Kwan, A.H.Y, Sum, E.Y, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2002-06-30 | | Release date: | 2003-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4
EMBO J., 22, 2003
|
|
1OW5
 
 | |
1J3G
 
 | | Solution structure of Citrobacter Freundii AmpD | | Descriptor: | AmpD protein, ZINC ION | | Authors: | Liepinsh, E, Genereux, C, Dehareng, D, Joris, B, Otting, G. | | Deposit date: | 2003-01-31 | | Release date: | 2003-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Citrobacter freundii AmpD, Comparison with Bacteriophage T7 Lysozyme
and Homology with PGRP Domains
J.Mol.Biol., 327, 2003
|
|
1MM0
 
 | | Solution structure of termicin, an antimicrobial peptide from the termite Pseudacanthotermes spiniger | | Descriptor: | Termicin | | Authors: | Da Silva, P, Jouvensal, L, Lamberty, M, Bulet, P, Caille, A, Vovelle, F. | | Deposit date: | 2002-09-02 | | Release date: | 2003-05-13 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of termicin, an antimicrobial peptide from the termite Pseudacanthotermes spiniger
PROTEIN SCI., 12, 2003
|
|
2PDX
 
 | | Human aldose reductase double mutant S302R-C303D complexed with zopolrestat. | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-04-01 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
2PDL
 
 | |
2C2S
 
 | | Human Dihydrofolate Reductase Complexed With NADPH and 2,4-Diamino-5-(1-o-carboranylmethyl)-6-methylpyrimidine, A novel boron containing, nonclassical Antifolate | | Descriptor: | 2,4-DIAMINO-5-(1-O-CARBORANYLMETHYL)-6-METHYLPYRIMIDINE, DIHYDROFOLATE REDUCTASE, GLYCEROL, ... | | Authors: | Leung, A.K.W, Reynolds, R.C, Riordan, J.M, Borhani, D.W. | | Deposit date: | 2005-09-29 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel Boron-Containing, Nonclassical Antifolates: Synthesis and Preliminary Biological and Structural Evaluation.
J.Med.Chem., 50, 2007
|
|
1TBN
 
 | | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN KINASE C, GAMMA TYPE, ZINC ION | | Authors: | Xu, R.X, Pawelczyk, T, Xia, T, Brown, S.C. | | Deposit date: | 1997-04-15 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions.
Biochemistry, 36, 1997
|
|
1I4V
 
 | |
2KBF
 
 | |
1IG0
 
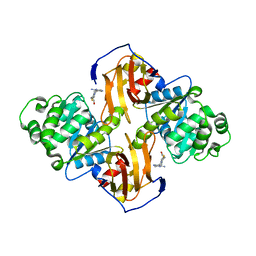 | | Crystal Structure of yeast Thiamin Pyrophosphokinase | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Thiamin pyrophosphokinase | | Authors: | Baker, L.-J, Dorocke, J.A, Harris, R.A, Timm, D.E. | | Deposit date: | 2001-04-16 | | Release date: | 2001-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of yeast thiamin pyrophosphokinase.
Structure, 9, 2001
|
|
