7JN9
 
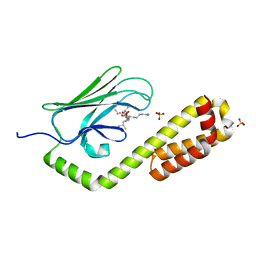 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide QEHTGSQLRIAAYGP | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4QMO
 
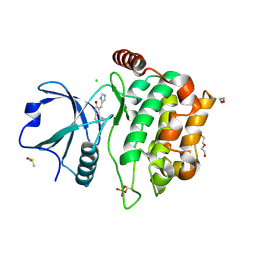 | | MST3 IN COMPLEX WITH Imidazolo-oxindole PKR inhibitor C16 | | Descriptor: | (8Z)-8-(1H-imidazol-5-ylmethylidene)-6,8-dihydro-7H-[1,3]thiazolo[5,4-e]indol-7-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Olesen, S.H, Watts, C, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2014-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Discovery of Diverse Small-Molecule Inhibitors of Mammalian Sterile20-like Kinase 3 (MST3).
Chemmedchem, 11, 2016
|
|
4O5G
 
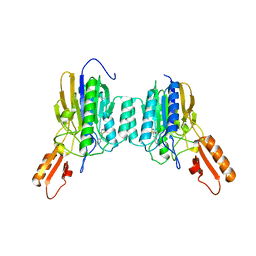 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{E})-5-[(4-aminophenyl)methylidene]-2-azanylidene-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
4CDU
 
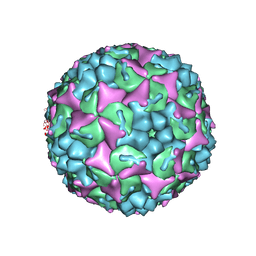 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-06 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
5DGS
 
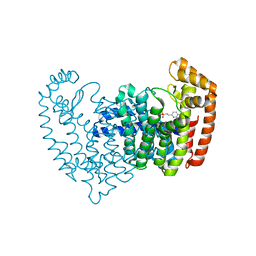 | |
4CEY
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor NLD | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
2P4B
 
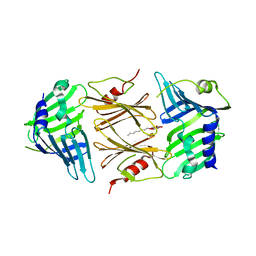 | | Crystal structure of E.coli RseB | | Descriptor: | Sigma-E factor regulatory protein rseB, octyl beta-D-glucopyranoside | | Authors: | Kim, D.Y, Kim, K.K. | | Deposit date: | 2007-03-12 | | Release date: | 2007-05-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of RseB and a model of its binding mode to RseA
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PLK
 
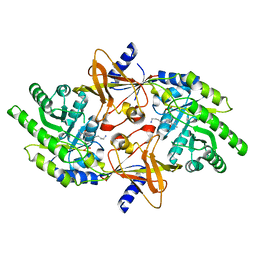 | |
1PYT
 
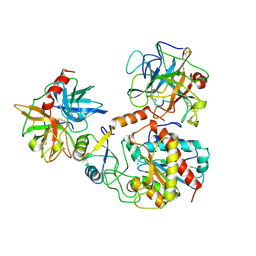 | | TERNARY COMPLEX OF PROCARBOXYPEPTIDASE A, PROPROTEINASE E, AND CHYMOTRYPSINOGEN C | | Descriptor: | CALCIUM ION, CHYMOTRYPSINOGEN C, PROCARBOXYPEPTIDASE A, ... | | Authors: | Gomis-Ruth, F.X, Gomez, M, Bode, W, Huber, R, Aviles, F.X. | | Deposit date: | 1995-06-21 | | Release date: | 1997-01-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The three-dimensional structure of the native ternary complex of bovine pancreatic procarboxypeptidase A with proproteinase E and chymotrypsinogen C.
EMBO J., 14, 1995
|
|
2A7U
 
 | | NMR solution structure of the E.coli F-ATPase delta subunit N-terminal domain in complex with alpha subunit N-terminal 22 residues | | Descriptor: | ATP synthase alpha chain, ATP synthase delta chain | | Authors: | Wilkens, S, Borchardt, D, Weber, J, Senior, A.E. | | Deposit date: | 2005-07-06 | | Release date: | 2005-10-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Interaction of the delta and alpha Subunits of the Escherichia coli F(1)F(0)-ATP Synthase by NMR Spectroscopy
Biochemistry, 44, 2005
|
|
1PS6
 
 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-HYDROXY-L-THREONINE-5-MONOPHOSPHATE, 4-hydroxythreonine-4-phosphate dehydrogenase, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M. | | Deposit date: | 2003-06-20 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
4RZB
 
 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate, SOAKED WITH MERCURY | | Descriptor: | GLYCEROL, MERCURY (II) ION, N-[(E)-iminomethyl]-L-aspartic acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-12-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
1RDZ
 
 | | T-STATE STRUCTURE OF THE ARG 243 TO ALA MUTANT OF PIG KIDNEY FRUCTOSE 1,6-BISPHOSPHATASE EXPRESSED IN E. COLI | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Stec, B, Abraham, R, Giroux, E, Kantrowitz, E.R. | | Deposit date: | 1996-05-17 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the active site mutant (Arg-243-->Ala) in the T and R allosteric states of pig kidney fructose-1,6-bisphosphatase expressed in Escherichia coli.
Protein Sci., 5, 1996
|
|
1RDY
 
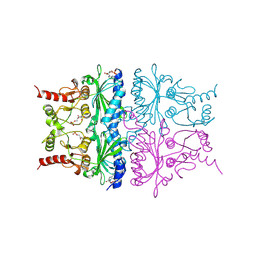 | | T-STATE STRUCTURE OF THE ARG 243 TO ALA MUTANT OF PIG KIDNEY FRUCTOSE 1,6-BISPHOSPHATASE EXPRESSED IN E. COLI | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Stec, B, Abraham, R, Giroux, E, Kantrowitz, E.R. | | Deposit date: | 1996-05-17 | | Release date: | 1997-01-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the active site mutant (Arg-243-->Ala) in the T and R allosteric states of pig kidney fructose-1,6-bisphosphatase expressed in Escherichia coli.
Protein Sci., 5, 1996
|
|
2KOR
 
 | |
7KA1
 
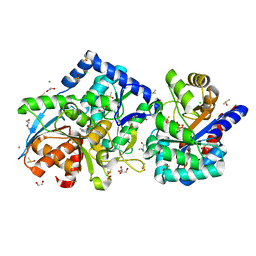 | | 1.60 Angstrom resolution crystal structure of the beta-Q114A mutant Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and cesium ion at the metal coordination site | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.60 Angstrom resolution crystal structure of the beta-Q114A mutant Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and cesium ion at the metal coordination site.
To be Published
|
|
1QB5
 
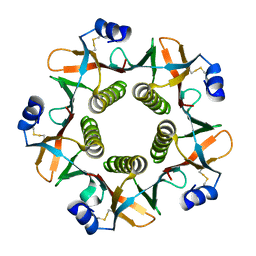 | |
2GNK
 
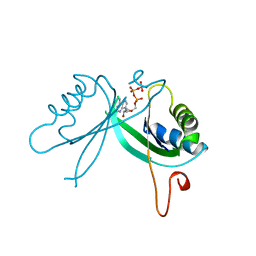 | | GLNK, A SIGNAL PROTEIN FROM E. COLI | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PROTEIN (NITROGEN REGULATORY PROTEIN) | | Authors: | Xu, Y, Cheah, E, Carr, P.D, van Heeswijk, W.C, Westerhoff, H.V, Vasudevan, S.G, Ollis, D.L. | | Deposit date: | 1998-07-14 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GlnK, a PII-homologue: structure reveals ATP binding site and indicates how the T-loops may be involved in molecular recognition.
J.Mol.Biol., 282, 1998
|
|
4CEW
 
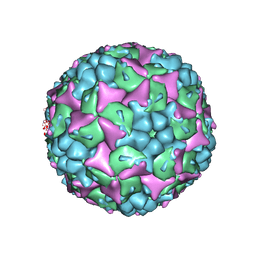 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor ALD | | Descriptor: | 4-[3-[(3s)-5-[4-[(e)-ethoxyiminomethyl]phenoxy]-3-methyl-pentyl]-2-oxidanylidene-imidazolidin-1-yl]pyridine-2-carboxamide, VP1, VP2, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5D6Q
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-{4-[(E)-2-(pyridin-3-yl)ethenyl]-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl}urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
1MUG
 
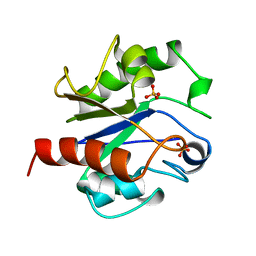 | | G:T/U MISMATCH-SPECIFIC DNA GLYCOSYLASE FROM E.COLI | | Descriptor: | PROTEIN (G:T/U SPECIFIC DNA GLYCOSYLASE), SULFATE ION | | Authors: | Barrett, T.E, Savva, R, Panayotou, G, Brown, T, Barlow, T, Jiricny, J, Pearl, L.H. | | Deposit date: | 1998-07-10 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions.
Cell(Cambridge,Mass.), 92, 1998
|
|
1RDX
 
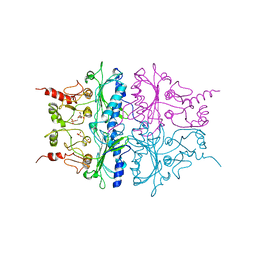 | | R-STATE STRUCTURE OF THE ARG 243 TO ALA MUTANT OF PIG KIDNEY FRUCTOSE 1,6-BISPHOSPHATASE EXPRESSED IN E. COLI | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Stec, B, Abraham, R, Giroux, E, Kantrowitz, E.R. | | Deposit date: | 1996-05-17 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the active site mutant (Arg-243-->Ala) in the T and R allosteric states of pig kidney fructose-1,6-bisphosphatase expressed in Escherichia coli.
Protein Sci., 5, 1996
|
|
1IE3
 
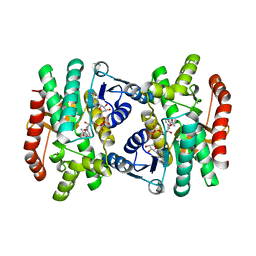 | | CRYSTAL STRUCTURE OF R153C E. COLI MALATE DEHYDROGENASE | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID | | Authors: | Bell, J.K, Yennawar, H.P, Wright, S.K, Thompson, J.R, Viola, R.E, Banaszak, L.J. | | Deposit date: | 2001-04-05 | | Release date: | 2001-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analyses of a Malate Dehydrogenase with a Variable Active Site
J.Biol.Chem., 276, 2001
|
|
2QVF
 
 | | mouse E-cadherin domains 1,2 | | Descriptor: | CALCIUM ION, Epithelial-cadherin (E-cadherin) (Uvomorulin) (Cadherin-1) (ARC-1) (CD324 antigen) | | Authors: | Carroll, K.J. | | Deposit date: | 2007-08-08 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | E-cadherin domains 1,2
To be Published
|
|
3DHX
 
 | | Crystal structure of isolated C2 domain of the methionine uptake transporter | | Descriptor: | IODIDE ION, Methionine import ATP-binding protein metN | | Authors: | Johnson, E, Kaiser, J.T, Lee, A.T, Rees, D.C. | | Deposit date: | 2008-06-18 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation.
Science, 321, 2008
|
|
